CEP63/FXR1/YAP1 axis in colorectal cancer progression
Published in Cancer
Centrosome is vital to maintain the precise separation of chromosomes and ensure chromosome stability1. Centrosomal proteins (CEPs) are dominant components of centrosome. Abnormal expression or mutation of CEPs may induce chromosomal instability and tumorigenesis2. CEP63, a member of CEPs, locates at the proximal end of the centrioles to promote de novo centriole biogenesis3. CEP63 deficiency and mutation can impede centrosome duplication and lead to asymmetric division4-5. However, there are few reports on the function and specific mechanism of CEP63 in tumors. Colorectal cancer (CRC) is a malignant disease with extremely high morbidity and mortality6, and most of sporadic CRC are characterized with chromosomal instability. But whether and how CEPs are involved in the pathogenesis remains unclear. In this study we explored the role of CEP63 in CRC development through in vitro and in vivo functional and mechanism assays.
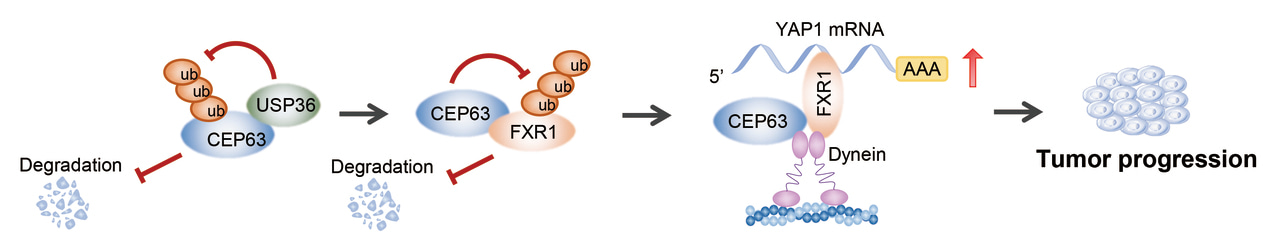
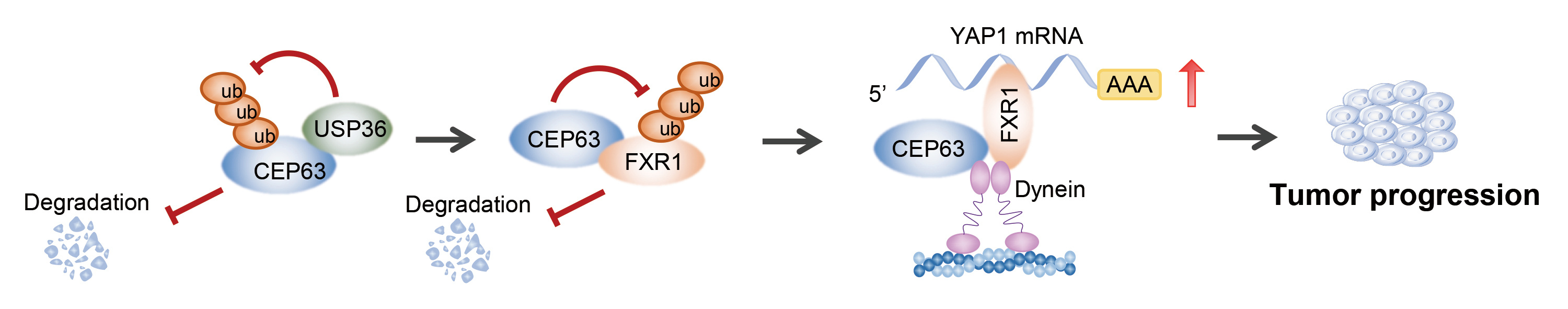
Firstly, we found that CEP63 protein was elevated in CRC tissues and correlated with a poor prognosis. And a series of in vivo and in vitro functional assays confirmed that CEP63 promoted CRC cell proliferation and tumor growth. Interestingly, we observed that the protein level and mRNA level of CEP63 in CRC were inconsistent, which indicated that the upregulation of CEP63 in CRC might mainly due to its protein stability. Through immunoprecipitation-mass Spectrometry (IP-MS) and ubiquitination-related assays, we verified that USP36 inhibited CEP63 protein degradation via reducing its K48-dependent ubiquitination modification.
Next, we wondered the underlying mechanism of CEP63 affecting the biological phenotype of CRC cells. Through transcriptome sequencing and verification, we identified that CEP63 promoted CRC stem cell-like phenotype by enhancing YAP1 expression. Subsequently, we revealed that CEP63 could bind multiple RNA binding protein (RBP) through IP-MS analysis and confirmed that one of the RBPs, FXR1, could mediate the regulation of CEP63 on the stability of YAP1 mRNA. Furthermore, we confirmed that CEP63 stabilized FXR1 protein by binding with the KH domain of FXR1 and inhibiting its K63-dependent ubiquitination degradation. In addition, we found that both CEP63 and FXR1 could interact with dynein components, which was, at least partly, mediated in CEP63/FXR1/YAP1 pathway. Considering CEP63 binding with multiple other RBPs, the data suggested that CEP63 exerted function as a common partner of RBP to regulate the destiny of RNA. Therefore, we here proposed CEP63/RBPs/RNA axis and identified novel molecular function of CEP63. But the precise mechanism needed further investigation.
In conclusion, our study demonstrated the versatility of CEP63 in physiology and pathology besides its canonical role as a structural component in centrosome. We discovered biological function and molecular model of CEP63 in signal transduction, RBPs regulation and RNA procession.
References
- Conduit PT, Wainman A, Raff JW. Centrosome function and assembly in animal cells. Nature Reviews Molecular Cell Biology. 2015;16:611-624.
- Kumar A, Rajendran V, Sethumadhavan R, Purohit R. CEP proteins: the knights of centrosome dynasty. Protoplasma. 2013;250:965-983.
- Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, et al. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr Biol. 2013;23:265-270.
- Sir J, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, et al. A primary microcephaly protein complex forms a ring around parental centrioles. Nature Genetics. 2011;43:1147-1153.
- Smith E, Dejsuphong D, Balestrini A, Hampel M, Lenz C, Takeda S, et al. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nature Cell Biology. 2009;11:278-285.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249.
Follow the Topic
-
Oncogene

This journal aims to make substantial advances in our knowledge of processes that contribute to cancer by publishing outstanding research.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in