Chemo-mechanical forces modulate the topology dynamics of mesoscale DNA assemblies
Published in Chemistry
The research landscapes
Harvesting principles of base pairing in nucleic acids, researchers have coerced abundant biological molecules such as DNA or RNA to fold into 2D and 3D nano- and macro-structures. To make the folding process much simpler and precise, scientists cleverly compiled computer programs to design and visualize folded nano-structures. Research groups in Kent State University (Mao), Mie University (Suzuki), and Tohoku University (Kawamata) synthesized DNA origami nanosprings using six-helix dsDNA bundles.
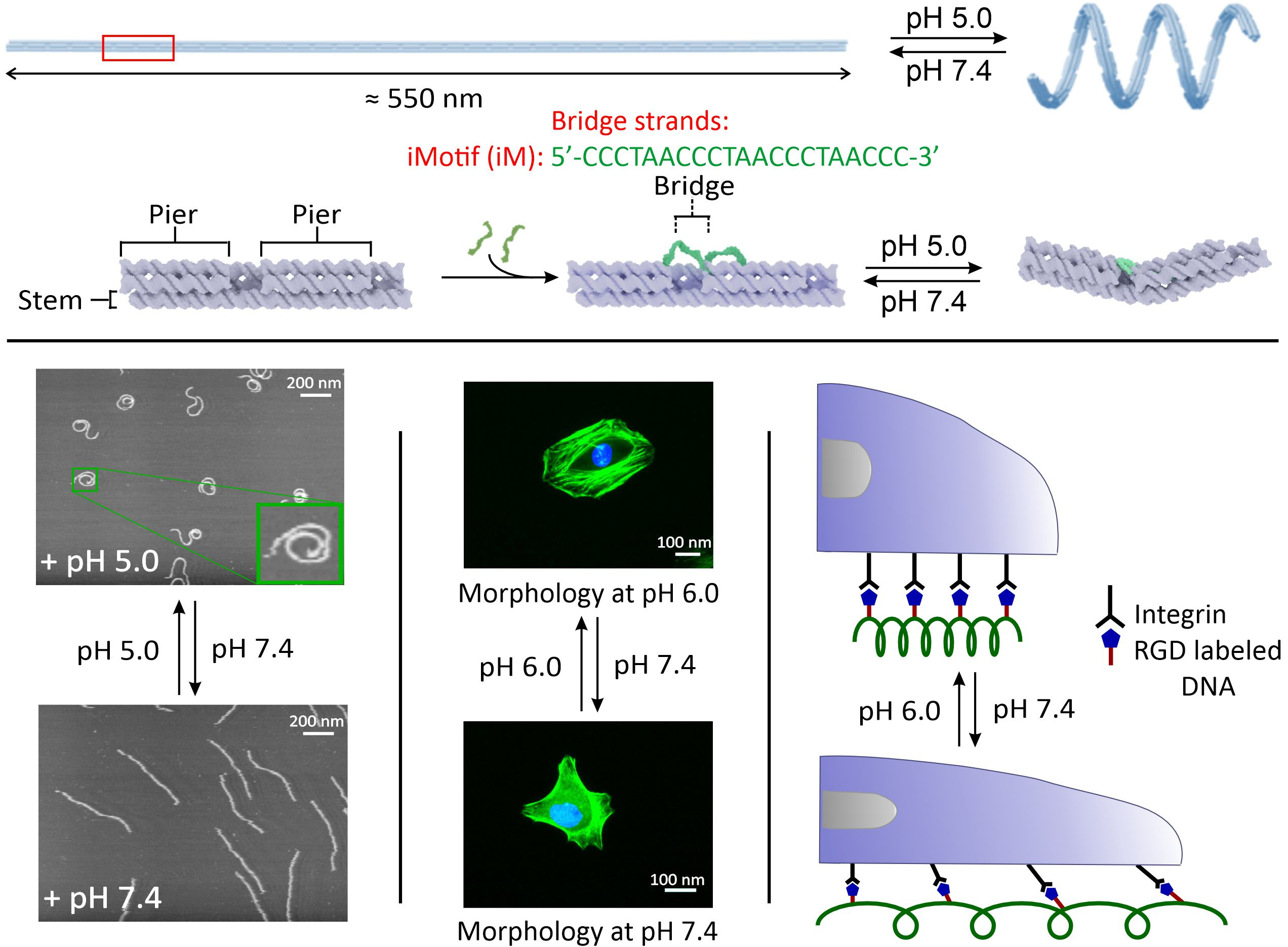
In the past two years, the research team was able to synthesize pH dependent nanospring origami by incorporating i-Motif structures in the nanospring backbone such that the origami demonstrated a linear conformation at neutral pH while a coiled form at acidic pH.1 Moreover, we modified the nanospring origami with arginyl-glycyl-aspartic acid (RGD) peptide, a ligand that binds to integrins on cell surface. This reversible conformation change was exploited to restrict the motion of cancer cells which experience acidic pH in extracellular medium (Figure 1). In addition, we also formulated a single-molecule method to characterize the mechanical properties of these nanosprings2, such as spring constant, kinetics, recoil and uncoil distances, and pitch of nanosprings.
Moving from nanoscale to mesoscale
In addition to elucidating nanoscopic properties of the DNA nanosprings, we also interrogated the mesoscale properties of these nanostructures. Since topological organizations can determine overall structure of a higher order assembly, it becomes relevant to divulge the structural-property relationship and the modulation factors behind the long-range, higher order arrangement of subunits in a mesoscale assembly. To that end, we synthesized two reversible forms of nanosprings such that the orientational flipping of their backbones could be achieved.3 We integrated several G-rich sequences (5’-TTAGGGTTAGGGTTAGGGTTAGGGTTA-3’) at the junctions of the nanospring backbone such that in presence of K+ ions, G-quadruplexes could be formed. The cumulative effect of all those folded quadruplexes provided a spring like coiled conformation. When a DNA sequence complimentary to the G-quadruplex, 5’-TAACCCTAACCCTAACCCTAACCCTAA-3’, or the anti-GQ DNA, was added to the nanospring backbone, a peculiar flipping of the backbone was observed, thereby giving off an impression of opposite coiling direction (Figure 2).
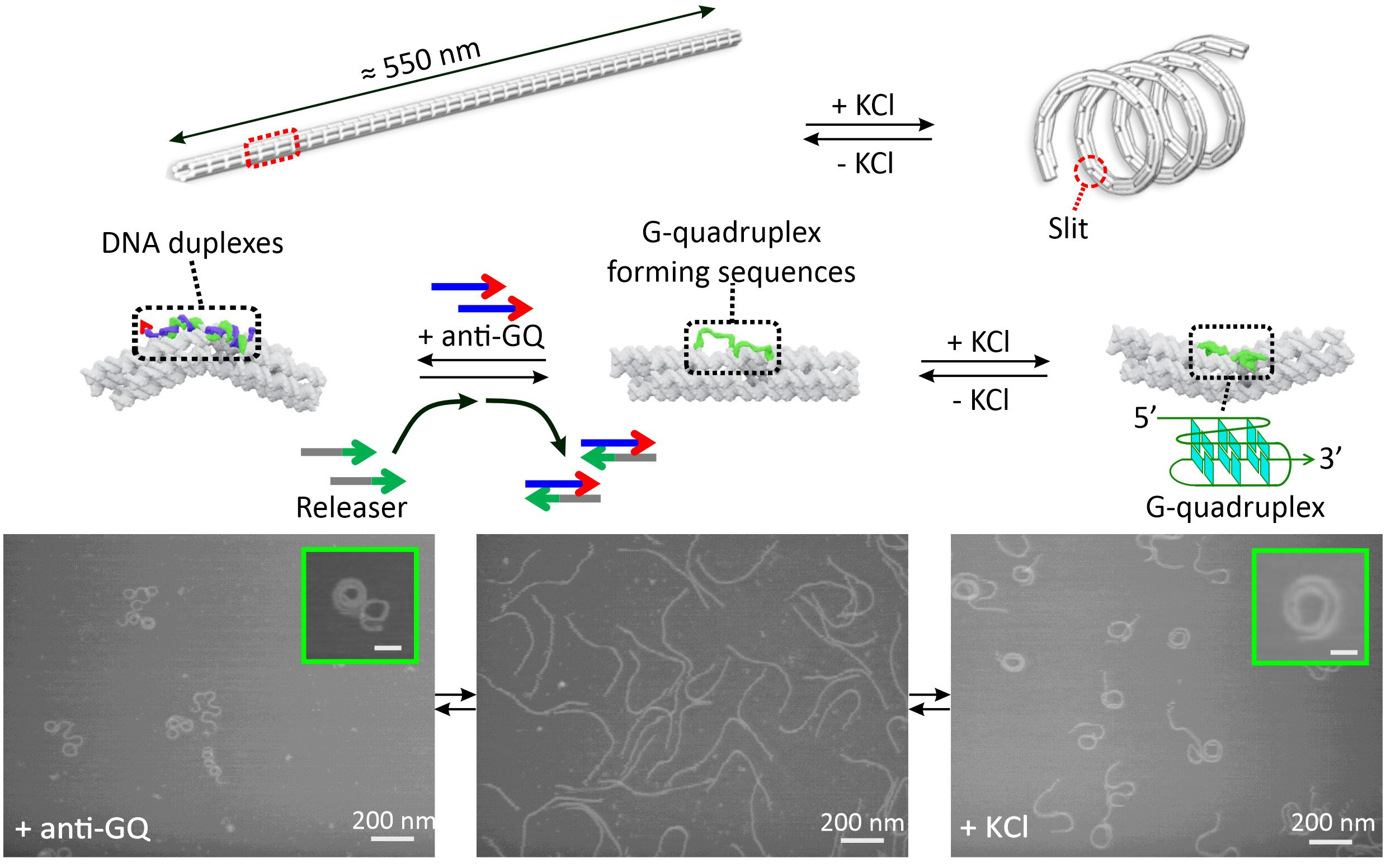
Figure 2. Reversible transformation of K+ and DNA responsive nanosprings.
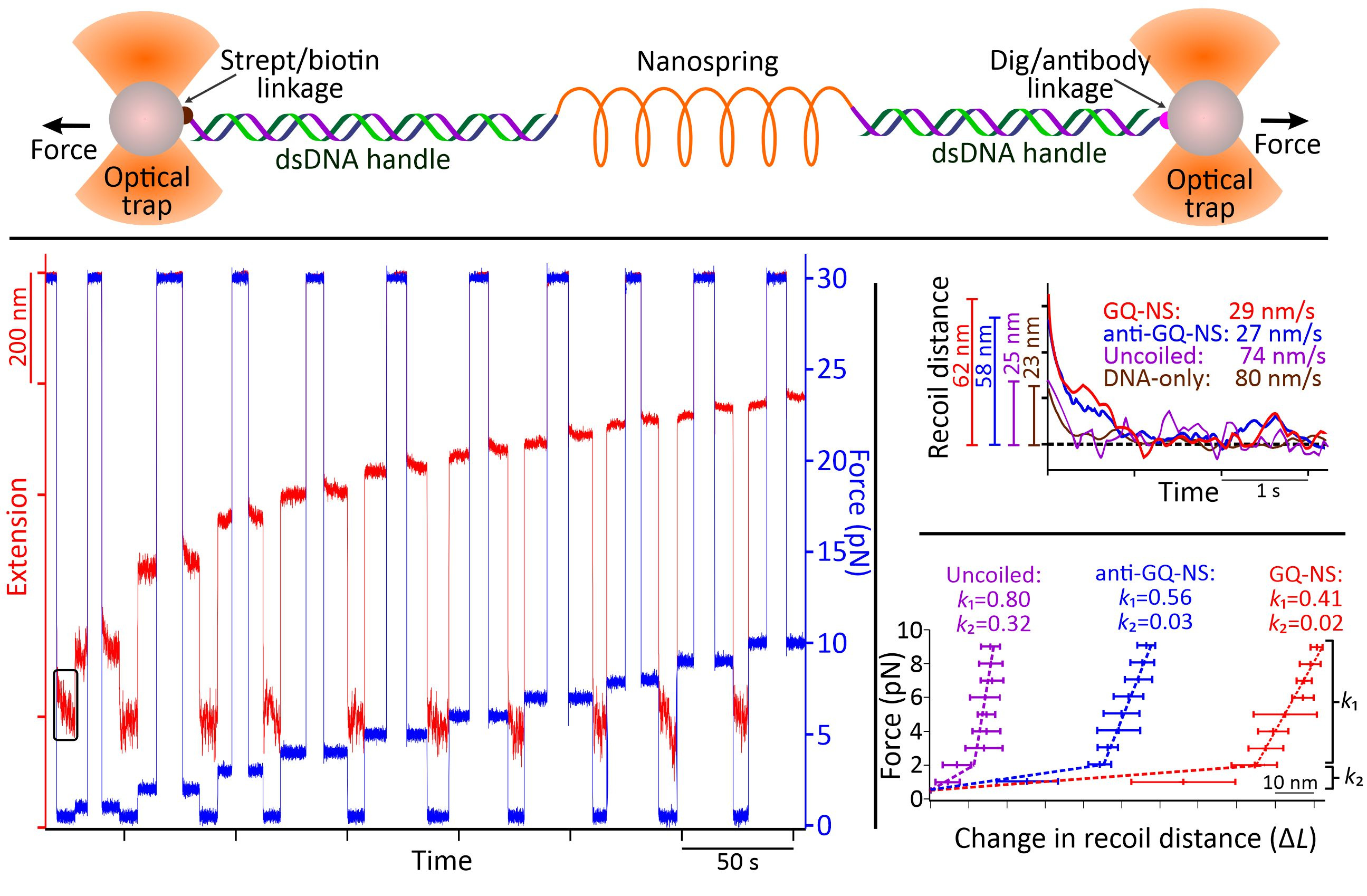
Through AFM image analysis, spirally coiled structures of nanosprings were observed in the presence of K+ or after incubation with anti-GQ DNA owing to the cumulative effect of the bending origami modules, while the origami without K+ or anti-GQ DNA showed a relaxed linear structure. Compared with the K+-induced nanospring (GQ-NS), the anti-GQ-DNA-incorporated nanospring (anti-GQ-NS) took a more coiled and compact structure as reflected in the statistical analyses of the AFM images. Measured values of the radius of the curvature and number of turns for anti-GQ-NS were 25.8 ± 2.8 nm (mean ± SD) and 3.5 ± 0.4, respectively, whereas those for GQ-NS were 43.9 ± 10.3 nm and 1.7 ± 0.4 nm. Using optical tweezers, we characterized the mechanical properties: the coiled length of GQ-NS was 189 ± 9 nm while that of anti-GQ-NS was 138 ± 3 nm. Likewise, the pitch length in 3D space was computed to be 110 ± 30 nm for GQ-NS and 39 ± 5 nm for anti-GQ-NS in 3D space. These structural features (shorter nanospring length and shorter pitches for anti-GQ-NS) at the resting state suggest a stronger spring constant for the anti-GQ-NS vs GQ-NS. We verified this hypothesis by performing a force jump assay in optical tweezers (Figure 3) and obtained recoiling rates for GQ-NS and anti-GQ-NS as 29 nm s-1 and 27 nm s-1 respectively. For the uncoiled nanospring (100 mM LiCl buffer without anti-GQ DNA), the recoiling rate was obtained as 74 nm s-1. The slower recoiling rates in nanosprings reflected the sluggish formation of the mesoscale coiling conformation. The longer recoiling distances for GQ-NS and anti-GQ-NS with respect to those observed in the uncoiled nanospring construct indicate mesoscale conformations have more dynamic ranges. In the uncoiled nanospring, the higher order mesoscale spring conformation is absent, therefore, the recoiling kinetics reflects inherent elastic behavior of the duplex DNA strand or bundles of dsDNA. Based on the recoiling distances at several forces, we used the Hooke’s law (force vs change in recoil distance) to obtain the spring constant. Interestingly, we observed two spring constants bifurcated at ≈2 pN, which is attributed to higher entropic contribution for coiling in the low force region (below 2 pN) and higher enthalpic contribution of the coiling above 2 pN. Above 2 pN, the spring constants were observed as 0.41 and 0.56 pN nm-1 for the GQ-NS and anti-GQ-NS, respectively, while below 2 pN, they were 0.02 and 0.03 pN nm-1, respectively. It is significant that anti-GQ-NS has higher spring constants than the GQ-NS, which agrees with the observation that anti-GQ-NS has smaller coil radius while shorter overall and pitch lengths at zero force.
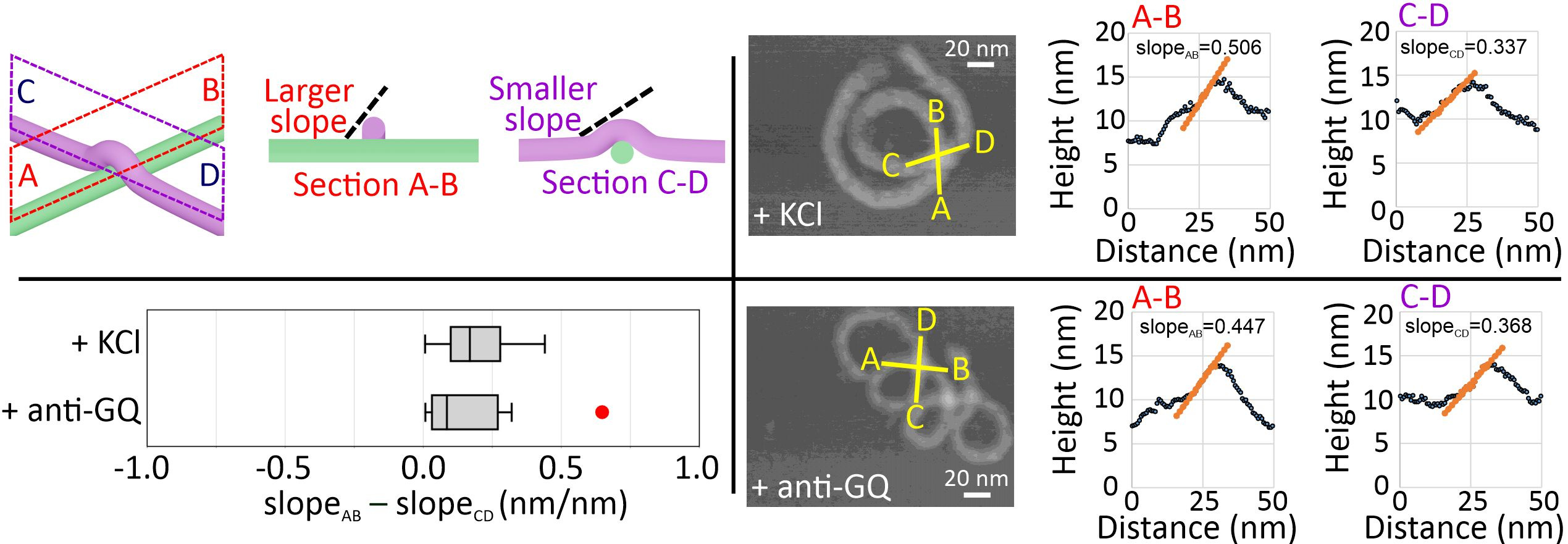
Next, we performed high-resolution AFM imaging to observe the coiling direction of the nanosprings by studying the intra-structure crossing-over points (Figure 4). Sectional profile analysis at the crossing-over point allowed us to judge which DNA bundle is lying underneath. This, in turn, enables us to distinguish the handedness of the nanospring. To determine such spatial features, we surveyed up-hill regions and compared the difference of the uphill slope values at the crossing-over point, slopeAB – slopeCD. A positive difference indicates a right-handed structure. In all our measurements, change in slopes for anti-GQ-NS and GQ-NS both showed positive values, suggesting both nanosprings are right-handed. Even though GQ-NS and anti-GQ-NS shared the right-handed helical chirality, opposite backbone orientations (i.e., backbone flipping) were observed. This suggests that an achiral backbone orientation stemmed from the linear chemo-mechanical force is not responsible for the higher order mesoscale chirality. Instead, the slightly overwinding helicity in the B-DNA based nanospring backbones may determine the nanospring chirality.
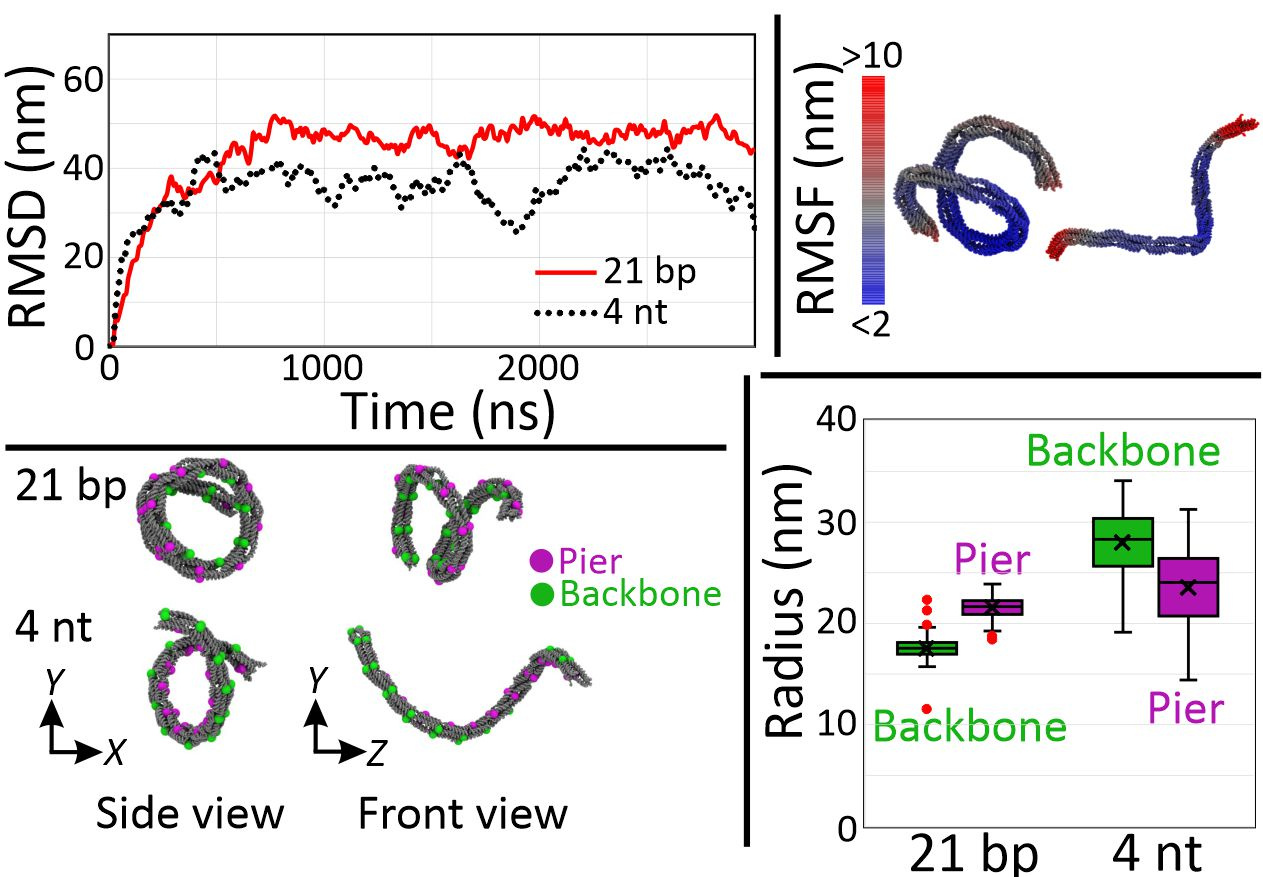
Finally, we performed coarse-grained molecular dynamics to confirm these higher order mesoscale structures of DNA nanosprings (Figure 5). To simulate a G-quadruplex structure, we used 4 nt poly-T linker (4T) given that both have similar end-to-end distances. Assuming the distance between the ends of G-quadruplex structure is about 2 to 3 nm and that between neighboring bases of single-stranded DNA is 0.68 nm, we simulated the structures as the 21 bp and the 4 nt linkers. From the plot of root mean square deviation (RMSD), we assumed that the structures were fully relaxed after 2 µs of simulation and decided to use the result of the last 1 µs for further analysis. Using 100 snapshots with 10 ns time interval of the last 1 µs, we computed the root mean square fluctuation (RMSF) and found that the structures were subject to large thermal fluctuations, indicating that the nanospring is a soft material. For the coiling arrangement, backbones appeared inside and outside for the cases of the 21 bp and 4 nt linker, respectively, which are consistent with experimental observations. In the case of the 21 bp linker, over-extended bridges push the piers apart, resulting in the inside backbones. On the other hand, tension from the short 4 nt linker pulls the piers together to make a coil with its backbone inside out. Finally, the difference in radius that the 4 nt has bigger coils than the 21 bp linker structure agreed with the experiment.
Perspectives
Based on the observation from mechanical stability of these two secondary structures (G-quadruplex and duplex DNA), we concluded that linear chemo-mechanical force in the range of 40 to 60 pN, which can be generated via formation/unfolding of biomolecular structures, is sufficient to control achiral higher order mesoscale structures such as DNA nanosprings. We anticipate these results not only provide insights on the origin of chiral helicity in mesoscale assemblies, but also lead to new, topologically based caging/uncaging strategies that are reversible in solutions. The topology-based backbone flipping could pave a way for delivering druggable compounds under environmental cues.
References
1 Karna, D. et al. Chemo-Mechanical Modulation of Cell Motions Using DNA Nanosprings. Bioconjugate Chemistry 32, 311-317, doi:10.1021/acs.bioconjchem.0c00674 (2021).
2 Karna, D., Pan, W., Pandey, S., Suzuki, Y. & Mao, H. Mechanochemical properties of DNA origami nanosprings revealed by force jumps in optical tweezers. Nanoscale 13, 8425-8430, doi:10.1039/d0nr08605c (2021).
3 Karna, D. et al. Chemo-mechanical forces modulate the topology dynamics of mesoscale DNA assemblies. Nature Communications 14, 6459 (2023).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in