Choosing the “best” split fluorescent proteins for your applications
Published in Protocols & Methods

Ever since our first paper on improved split FPs got published on Nature Communications in 2017, my advisor Professor Bo Huang and I consistently received emails asking how to choose the “best” split FPs for their specific needs. I ended up spending most of my “incubation time” (waiting time during multiple rounds of staining and washes) answering these questions point-by-point, as careful as I was writing a reply letter to an experienced peer reviewer.
This September, our new paper focused on split sfCherry3 and its application in multicolor visualization of neuronal synapses is out on Communications Biology. Similar questions started to popped up in my inbox again. I decide to take this chance to summarize my experience in developing split FP1-10/11, to describe some properties based on my observations, and hopefully to answer your question regarding the “best choice”.
Is split mNeonGreen2 the brightest among all?
The “brightness” is a vague claim depends on the cellular environment. If you want to endogenously tag a well-expressed protein in the cytosol through CRISPR knocking in the FP11, the simple answer is “Yes”. In the single-molecule level (when the complementation process has finished), the brightness of mNeonGreen21–10/11 is much higher than GFP1–10/11. However, in the bulk level, the complementation efficiency between the FP1-10 and FP11 fragments can be largely variable, with GFP1–10/11 being almost 100%, while mNeonGreen21–10/11 is less than 50%. In this case, if you cannot push the complementation to saturation by over-expressing the FP1-10 in cytosol (or any specific cellular compartment), using split GFP might actually give you better images. As for all red-colored split FPs, they’re relatively dimmer. Therefore, unless your green channel has been taken, start from mNeonGreen21–10/11.
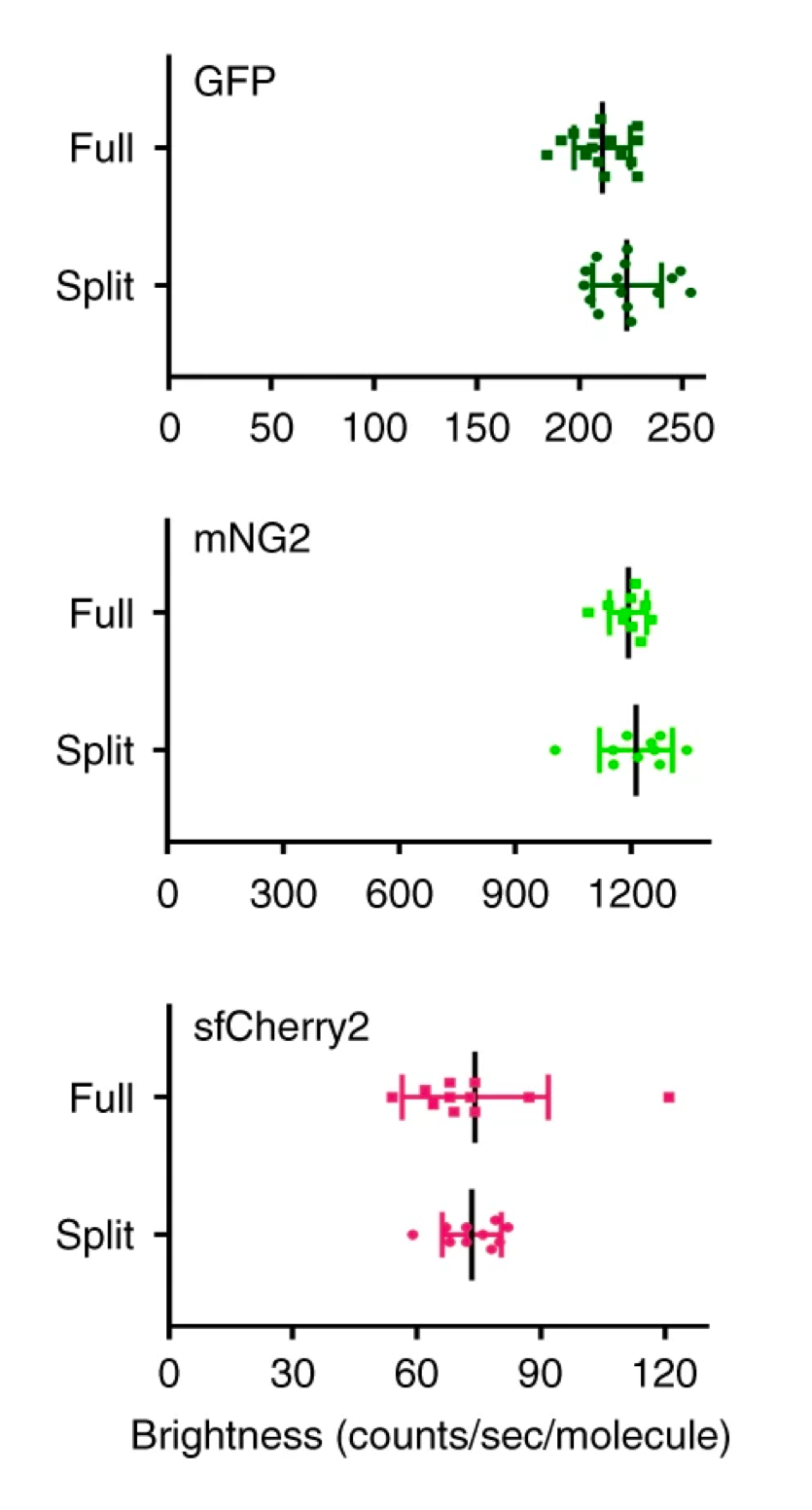
Figure 1: Single-molecule brightness measurement of split FPs and their full-length counterparts, demonstrating there is no significant different between the split and the full-length.
Any performance difference between split sfCherry3C VS sfCherry3V?
We reported two split sfCherry3 variants in this work and we tried to avoid ranking one over another when writing the manuscript. The split sfCherry3C was the best mutant coming out from random mutagenesis screening, while the sfCherry3V has been intentionally designed through introducing a L83W mutation inspired by a circular permutated mCherry variant (with enhanced protein folding efficiency). Our initial intention is to enhance sfCherry3C1–10/11 complementation efficiency so that the overall brightness would be higher. Unluckily, we did not observe a general fluorescence signal boost across multiple protein targets labeled through endogenous knock-in. But we do obtain clearer and cleaner ER (endoplasmic reticulum) structures using sfCherry3V1-10. Our explanation is the mother protein sfCherry has such a stable backbone that is resistant to lysosomal degradation. When labeling proteins involved in the endosomal pathway which normally ends in lysosomes, they tend to be aggregated inside and keep shining fluorescence. These granule-like aggregations mask the beautiful structure of the original targets (such as ER tubules). The L83W mutation might potentially destabilize the rigid backbone and make it more vulnerable to lysosomal degradation. One evidence is we haven’t seen any granule-like aggregations in knock-in cell lines after switching to sfCherry3V1–10/11. In short, if your protein of interest will be go through the endosomal pathway, try sfCherry3V first; otherwise sfCherry3C is generally a good choice.
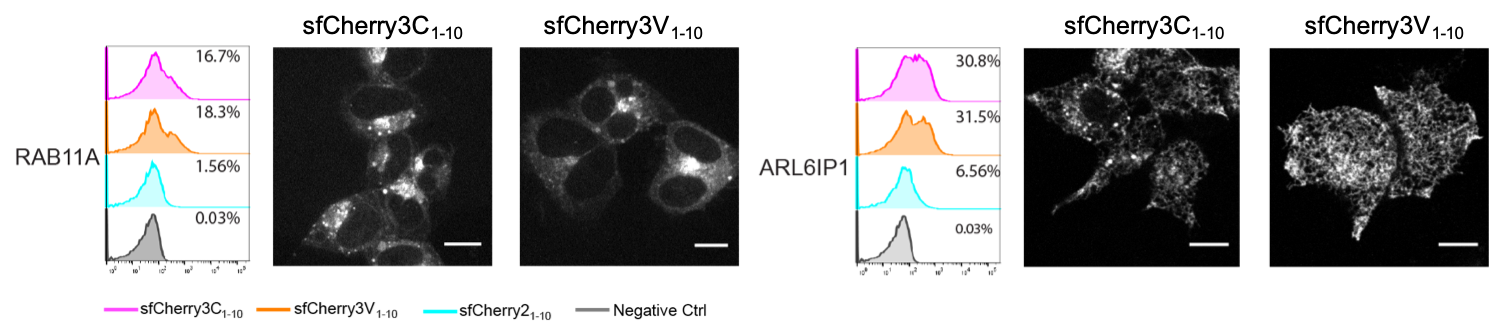
Figure 2: Endogenous protein labeling in HEK 293T cells using sfCherry3 variants. RAB11A and ARL6IP1 are both involved in the endosomal pathway. The microscopy images from sfCherry3V is crispy and have less punctate structures.
Will tandem FP11 tag always give proportional signal amplification?
That is NOT true. For split FP having ~100% complementation, we observed a fluorescence signal improvement linearly to the copy number of FP11 tags (the only case is in GFP1–10/11). However, considering the incomplete complementation, we did not achieve proportional signal boost by using mNeonGreen211, potentially limited by the expression level of mNeonGreen21–10 fragment. We have another enhanced version called mNeonGreen31–10/11 in pipeline, which demonstrates improved signal amplification in tandem tags. The alternative way for increasing the overall labeling brightness is to assist complementation through SpyTag/SpyCatcher interaction.
To conclude, there is no unified standard to elect the best split FP for all biological applications. With such unique and extensive applications of split fluorescent protein tool, it’s safe for us to expect the evolution wheel guided by molecular engineers will never stop.
References:
Feng, S., Varshney, A., Villa, D.C., Modavi, C., Kohler, J., Farah, F., Zhou, S., Ali, N., Müller, J.D., Van Hoven, M.K. and Huang, B., 2019. Bright split red fluorescent proteins for the visualization of endogenous proteins and synapses. Communications biology, 2.
Feng, Siyu, Sayaka Sekine, Veronica Pessino, Han Li, Manuel D. Leonetti, and Bo Huang. "Improved split fluorescent proteins for endogenous protein labeling." Nature communications 8, no. 1 (2017): 370.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
I love the FAQ format! I'm sure this will be really useful for other researchers.