Climate change vulnerability of forest trees in the age of genomics
Published in Ecology & Evolution

Accelerated climate change is one of the greatest current threats to biodiversity but an understanding of how taxa will respond to future climate change is mostly lacking. Efforts to predict future species response to climate change mostly assume that the species is static and hence ignore that species comprise genetically diverse individuals. However, intraspecific adaptive genetic variation could provide the raw material necessary to tolerate or adapt to environmental change. Therefore, it is critical to incorporate such information in the assessment of species vulnerability and conservation management under future climate change.

Forest trees play a leading role in the global carbon cycle and, along with being the most efficient carbon sink, forest trees will play an increasingly important role in combating climate change and global warming. However, forest trees are characterized by long lifespans, large body sizes and often have long generation times that make them particularly vulnerable to maladaptation under altered climatic scenarios. Some threats due to climate change may even happen within the lifetime of single individuals. Temperate forests in East Asia have high biodiversity, varied ecosystems, and house substantial endemism. However, compared to other regions, temperate forests in this area still lack a comprehensive evaluations of potential climate change impacts. Our study focused on one of the dominant tree species in temperate deciduous forest in East Asia, Populus koreana, to decipher its past demographic history, to reveal the genetic basis of current climate adaptation, and lastly to predict the future vulnerability under climate change.
How to dissect and identify the genetic variation underlying plants’ adaptation to their environments?
To understand the genetic basis of environmental adaptation and to identify the variation underlying the adaptation of P. koreana, we first generate a de novo high-quality chromosome-scale reference genome for this species. We then utilize large-scale resequencing datasets from 24 natural populations to explore not only SNPs, but also to characterize the short insertion/deletions (Indels) and larger structural variations (SVs). The landscape genomic approaches facilitate a thorough identification of genetic variants that underpin climatic adaptation, and some of which are validated by our experimental functional tests. Overall, our results revealed that the local adaptation of P. koreana to the current variable environments evolved by small polygenic allele frequency shifts as found for many other species.
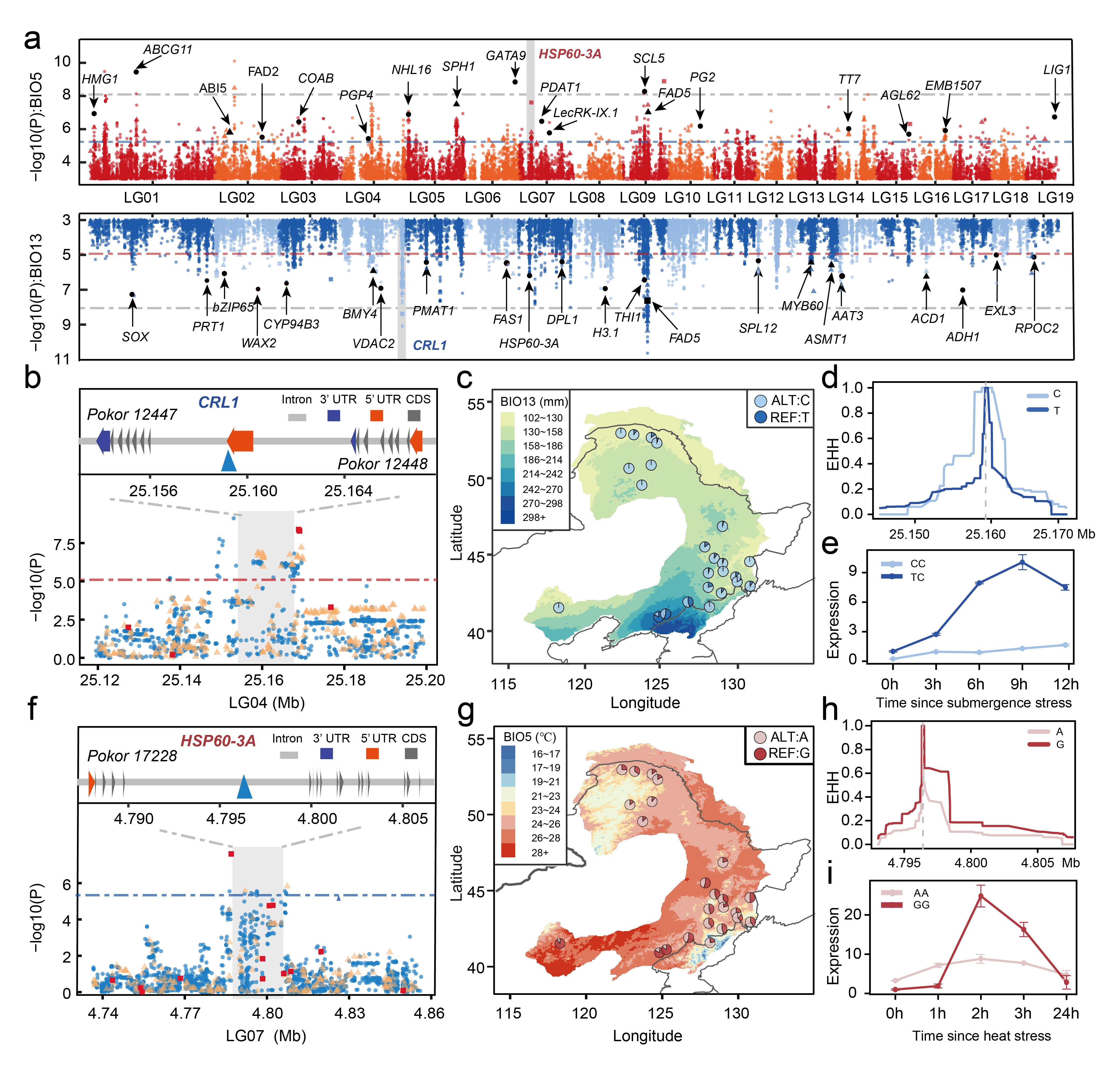
How to predict the responses of species and populations to rapid future climate change?
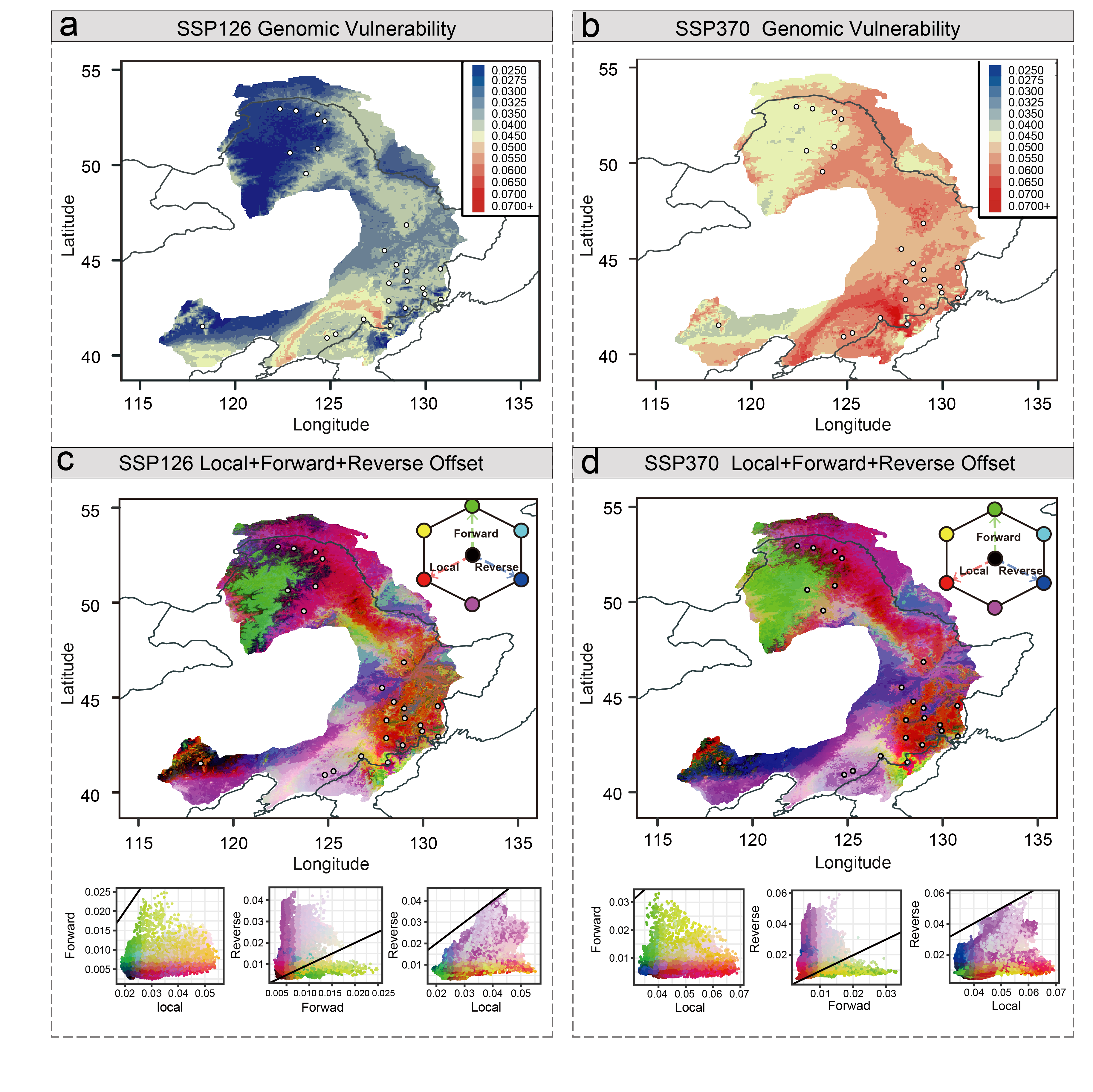
Based on the established contemporary genotype–environment relationships and the identified climate-associated genetic loci, we further predict how populations of P. koreana will respond to future climate change. We used various complementary approaches, including both single-locus-based (RONA), multi-locus-based (GF) as well as models that integrate migration into the analyses, to investigate the spatial pattern of maladaptation across the range of P. koreanaunder future climate conditions. All the results showed that a set of populations located in the southeastern part of the current distribution range are most likely to be vulnerable in the future. These populations, therefore, are needed to be conserved with special management strategies not only because of their high genomic offset to future climate change but also because they contained many unique, climate-adaptive genetic resources. Taken together, our findings highlight the importance of integrating genomic and environmental data to predict adaptive capacity of a key forest to rapid climate change in the future, and also identify many candidate genes and variants that may be useful for future forest tree breeding with special aims.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Hi, Jing, Well Done!
It would be more interesting your field story and field photos here~~Field work is really fun, cheers~