ContScout: removing contamination from annotated genomes
Published in Ecology & Evolution, Protocols & Methods, and Genetics & Genomics
Technological improvements in high-throughput sequencing have led to an unprecedented increase in the number of sequenced genomes in public databases. Several large-scale sequencing projects are underway focusing on specific taxonomic groups such as insects, fungi, vertebrates or plants with the ultimate goal of capturing the entire biodiversity of the Earth. On the way to this goal an important question arises when we look at the published genomes: can we really believe the images that we see? (Fig. 1)
Image sources: Fliegende Blätter (1892), Jorge Rodríguez (Pixabay)
Draft genomes are by definition incomplete. They often lack information about regions that are difficult to sequence, assemble or annotate. On the other hand, published genomes may contain excess sequences that do not belong to the target organism but are results of contamination. If these sequences remain in the data set, they can affect all subsequent comparative genomic phylogenomic analysis and possibly lead to completely wrong conclusions as was the case with the infamous tardigrade genome1. There, foreign sequences introduced by massive bacterial contamination were mistaken for horizontal gene transfer between bacteria and eukaryote.
In our article, recently published in Nature Communications, we present a tool called ContScout which was developed to identify and remove contamination from annotated draft genomes. The key feature that distinguishes ContScout from other decontamination software is its ability to combine taxonomic information with gene locus information (Fig. 2).
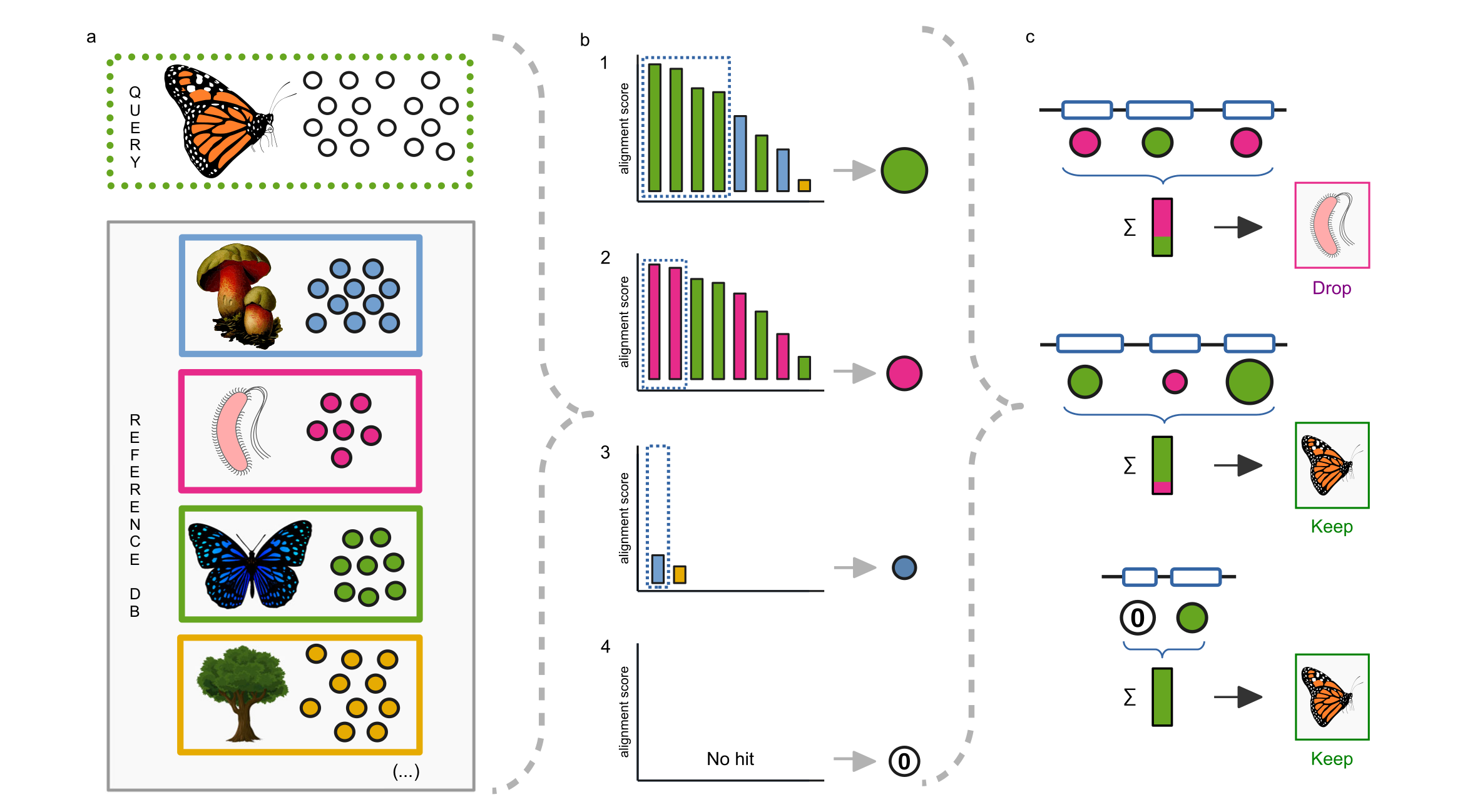
Using synthetic data and manually filtered sequences, we demonstrate that ContScout has an outstanding sensitivity and specificity, largely outperforming most taxon labeling / sequence decontaminating tools available to date (Conterminator, BASTA, MMSeqs and DIAMOND). By screening 844 published genomes representing all major clades of the eukaryotic tree of life, we show that contamination is widespread among them with some extreme cases containing thousands of foreign proteins. (Fig. 3)

Using a ubiquitous pyridoxal kinase protein family as an example, we show in detail how the presence of two mislabelled proteins can confound the inferred evolutionary history of the gene family by introducing 15 excess deletion events throughout the species and several gene gain events near the root of the tree. (Fig. 4)
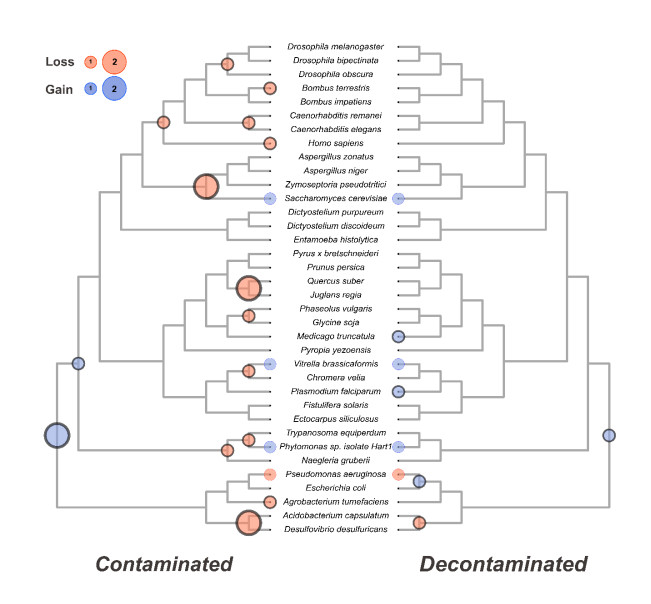
Using 36-genome demonstration data set and a leave-one out approach, we show that the confounding effect of contaminated genomes on ancestral gene content estimates is profound and cumulative. Finally, we challenge ContScout with a set of known HGT cases. We prove that the tool is able to distinguish HGT from contamination in most of the tested cases.
With our article, we aim to draw the attention of the scientific community on the prevalence of contamination among published genomes and show how unhandled contamination can bias phylogenomics studies. Furthermore, we present a user-friendly tool for the detection and removal of contamination in annotated genomes.
Separating closely related genomes from each other with ContScout posed the biggest challenge that we encountered during the development. This major improvement was unanimously suggested by two of the referees during the manuscript revision procedure. In order to enable this feature, the taxon calling module needed a complete redesigning and re-implementation that took nearly two months to complete. I firmly believe that this improvement is a game changer and hope that the tool will prove useful for the research community.
References:
1. Boothby TC et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A. (2015)
doi: 10.1073/pnas.1510461112.
2. Nagy LG et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat Commun. (2014)
doi: 10.1038/ncomms5471
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in