Counting Chromosomes: The Simple Idea That Unlocked the Rules Behind Centromere Evolution
Published in Ecology & Evolution and Cell & Molecular Biology
How many chromosomes do different species of budding yeast have? That was the question we were trying to answer last year to provide evolutionary context to our finding that baker's yeast needs at least five chromosomes to divide properly (Helsen et al. 2024). At the time, we never expected to learn about centromeres.
We started by scouring the literature for species with known chromosome numbers, but after trying to count and distinguish individual bands on one too many grainy pulse-field gels, we had to admit defeat. We concluded we'd only ever have an estimated chromosome number for a handful of species—far fewer than we needed to confidently say that, indeed, in nature too, different budding yeasts never have fewer than five chromosomes. Because of course, everything you see in the lab corresponds perfectly to what we see in nature... right?

Around the same time, the project also ran into another roadblock. To try and figure out why baker’s yeast can’t have fewer than five chromosomes, we had decided to evolve our strains in the lab, hoping to see what kind of solutions they would come up with to deal with having so few chromosomes. Only, it turned out that the way they deal with it is to diploidize. If you know anything about experimental evolution of baker’s yeast, you know that diploidization is generally thought to be the least exciting outcome of any experiment—these strains are often even excluded from further analyses. We were stuck and needed to take a step back.
The Naive, Simplistic Idea That Unlocked Another Mystery
To kill some time and hopefully come back to our original problem with a fresh perspective, we returned to our quest of counting chromosomes in different budding yeast species. This time, we had an idea. An extremely naive, simplistic idea, but an idea nonetheless: Why not count centromeres in genome assemblies? Supposedly, there should only be one centromere for each chromosome, and budding yeast centromeres should be small and non-repetitive enough to not span multiple contigs.
That’s how finding centromeres became one of my new favorite hobbies.
Finding new centromeres should really not have been as much fun as it was, but every time I was able to discover them I felt like a gold prospector discovering a nugget of gold or a detective finally finding that one final clue. My list of centromeres grew steadily, and with that, also the realisation that actually, these centromeres are incredibly interesting in and of themselves.

Centromeres are … peculiar. We know that they are one of the fastest-evolving parts of the genome, yet at the same time they have to maintain the absolutely essential function of correctly attaching chromosomes to the cell division machinery.
When I suddenly found genomes containing two different types of centromeres simultaneously, we knew this wasn't just about counting chromosomes anymore—this was going to be a story worth telling.
Uncovering the Evolutionary Rules
That's when our small counting project grew into a full-scale evolutionary investigation. Our work ultimately addresses a deep mystery in biology: how does the rapidly evolving centromere keep on working perfectly with the cell’s more conserved components of the division machinery?
What emerged was a fascinating picture. We found concrete proof that when a new centromere variant arises, it doesn't just instantly take over. Instead, we observe instances where two distinct types of centromeres exist within the population simultaneously. These variants then spread through a slow, step-by-step process of genetic drift and positive selection, changing their distribution one by one until one reaches fixation.
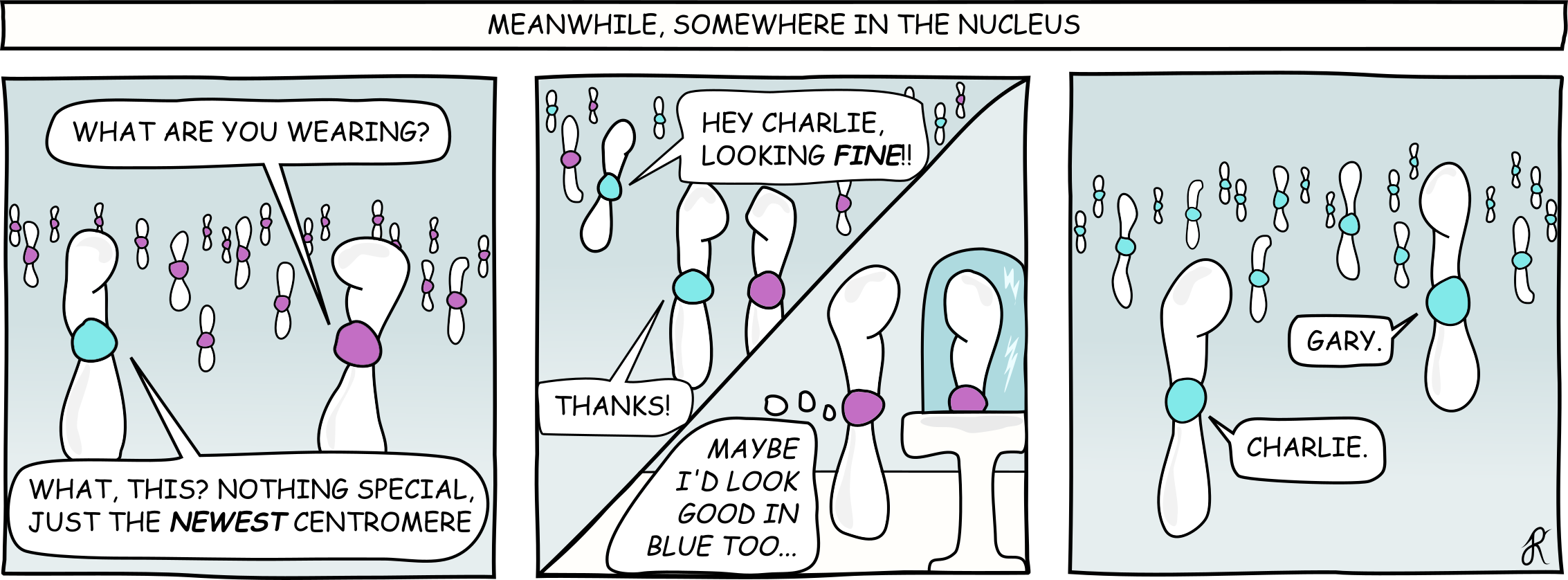
The key insight is that the cell's main division machinery acts like a strict quality control filter. Only those new centromere variants that can still latch onto the spindle (the cell's pulling mechanism) can survive and spread. If a variant is incompatible, it simply gets eliminated by natural selection. This finding gives us an unprecedented look at the evolutionary constraints—the rules—that govern how this crucial connection is maintained through evolution.
Why Now? The Role of Technology
We couldn't have done this ten years ago. Centromeres are notoriously messy, repetitive stretches of DNA. But thanks to the revolution in long-read sequencing technology, we can now get "telomere-to-telomere" genome assemblies. This means we can finally read through these complex regions without missing anything. This technological leap allows us to look beyond just the standard model organisms and study centromeres in hundreds of different species. Coupled with this, our work is one of the very first to directly test this coevolutionary dance between the centromere and the division machinery, making it a foundational reference for the field.
Ultimately, what started as a simple way of counting chromosomes became our key to unlocking the fundamental evolutionary rules governing centromere evolution.
And what about that first story, the one with the roadblocks? It turned out that the method we developed not only shed light on over a billion years of centromere history but also definitively answered our initial question: the smallest number of chromosomes in our dataset is indeed six. Very occasionally nature does behave as expected.
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcement



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in