
Background
The COVID-19 pandemic has generated, in the last two years, an explosion of data availability on the web, including SARS-CoV-2 viral sequences deposited from sequencing laboratories, research publications, and knowledge items spread around the unstructured web content. Many research organizations have studied the genome of the SARS-CoV-2 virus and a body of public resources have been published for monitoring its evolution.
While we experience an unprecedented richness of information in this domain, we also ascertained the lack of a systematic organization of such information. In our article recently published in Nature Scientific Data, we contribute to this issue by building a knowledge graph based on an abstract model called CoV2K, which allows us to represent both the data and the external knowledge that is being collected about SARS-CoV-2.

Our proposal
CoV2K contains areas with knowledge about SARS-CoV-2 (left part of the figure) and areas with data about the virus (right part of the figure). The 'Knowledge' part allows collecting information on variants and their names (produced either by organizations or by computational methods), their effects (such as their resistance to monoclonal antibodies, convalescent/vaccine sera, transmissibility, or virulence) as reported by literature evidence, and their composition (in terms of sets of mutations, which have specific positions through the structure of the viral genome/proteins). It then includes the peculiarities of mutations due to their original and alternative nucleotide or amino acid residues (i.e., amino acid changes residues features, including changes of polarity, hydrophobicity, or charge) and the definition of particular regions of the genome with given functions. The 'Data' part includes information about real collected sequences (with their describing metadata provided by laboratories and their mutations) in addition to epitopes tested for the virus and its hosts. Within and across the areas, entities are connected by relationships of various types, defining the conceptual connection between their concepts.
CoV2K provides a concise route map for understanding the types of information related to the virus and their connections; it serves as guidance to drive a process of data and knowledge integration that aggregates information from several current resources, harmonizing their content and overcoming incompleteness and inconsistency issues.
Building CoV2K
In building the CoV2K content, we have employed a classical data integration process driven by an abstract model, with pipelines for the integration and harmonization of different data silos (also shown in the figure above, as independent circles and rectangles at the borders of areas). For what concerns knowledge, we have chosen the information sources so that they are the most updated in the landscape of SARS-CoV-2-related knowledge and they provide a tight update schedule. Moreover, our ETL and harmonization pipelines for feeding CoV2K have been designed to allow easy extension of its content by future addition of data sources when these become available and are deemed trustworthy. We now integrated Variants and Effects information from several authoritative sources such as CoVariants.org, Public Health England, the COG-UK Mutation Explorer, ECDC, and several preprints or published papers deposited on bioRxiv, medRxiv, or PubMed. As structure and residues references we employed NCBI RefSeq, NCBI Structures, and UniProtKB. For what concerns data, CoV2K includes two large databases. We previously developed the ViruSurf database (http://gmql.eu/virusurf/), which at the time of publication 2022 includes around 5 million sequences from GenBank and COGUK with both nucleotide mutations and amino acid changes. Our pipelines reload and curate data regularly. We also include in CoV2K the Immune Epitope Database (IEDB, https://www.iedb.org/) containing about 6.5K epitopes defined for SARS-CoV-2. The current version of the CoV2K system is undergoing continuous updating of information. We are designing semi-supervised methods for extracting content from the CORD-19 literature corpus to continuously collect instances of knowledge-related entities.
How to use CoV2K
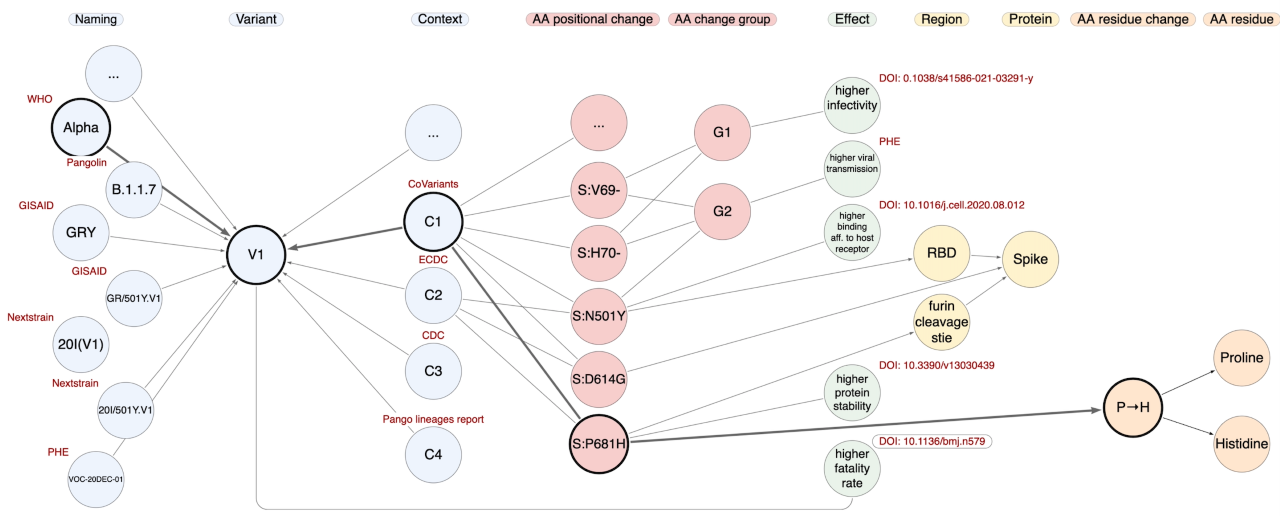
The silos integrated within CoV2K can be explored using a flexible API (http://gmql.eu/cov2k/api/) that navigates a graph. Through our API, users can address single entities or paths through their relationships, asking for information that regards SARS-CoV-2 knowledge. For instance, "What are the characteristics (Grantham distance and type) of the residue changes of the Alpha variant?" (some of the instances involved in this query are represented in the figure below), or "Which amino acid changes of VOC-20DEC-02 fall within the Receptor Binding Domain (RBD)?", or even "Which are the effects of the variants that include the Spike amino acid change D614G?". The most powerful use of CoV2K, however, can be made by connecting knowledge entities with data entities, e.g., "Which epitopes are impacted by amino acid changes with documented effects on the binding affinity to the host cell receptors?"
Towards FAIR systems and beyond
In the last year, we also built other systems that allow elaborate SARS-CoV-2 data (VirusViz, ViruClust, VariantHunter). It became apparent that connecting data with knowledge is of great importance. For example, when visualizing mutation distributions, it is important to connect specific mutational patterns to the area of the virus on which they insist, possibly knowing what functions that area brings. When observing specific mutations with an increasing or decreasing trend, it may be useful to compare them with the characterizing mutations of known lineages worldwide or to check the existence of studies on their effects on immunogenicity or disease treatment. In all these experiences, mastering the interplay between data and knowledge in SARS-CoV-2 has proven to be extremely useful. The linking of CoV2K concepts to our web resources is a step forward in promoting FAIR principles, as it facilitates – at the conceptual level – the interoperability between public data sources and open knowledge and – at the practical level – the creation of several future systems that will exploit the new possibilities allowed by interlinking data and knowledge.
Read the full, open-access article in Scientific Data at:
Alfonsi, T., Al Khalaf, R., Ceri, S. et al. CoV2K model, a comprehensive representation of SARS-CoV-2 knowledge and data interplay. Sci Data 9, 260 (2022). https://doi.org/10.1038/s41597-022-01348-9 (2022).
The CoV2K API is available at http://gmql.eu/cov2k/api/
Follow the Topic
Research Data
Research Communities > Community > Research Data
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
This Scientific Data Collection welcomes submissions of Data Descriptors associated with datasets for crop management, which are essential for optimising agricultural productivity, sustainability, and food security.
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
This Scientific Data collection aims to gather data descriptors on high-quality, reusable datasets relevant to the drug discovery and development process.
Publishing Model: Open Access
Deadline: Apr 22, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in