CRISPECTOR: A Better Tool for Measuring CRISPR-Cas-induced Adverse Activity
Published in Bioengineering & Biotechnology

Since its discovery, the CRISPR-Cas system has had a monumental impact on the field of gene-editing and the world of biotechnology as a whole. CRISPR technology allows researchers to modify gene function by editing genomes and altering DNA sequences. Its many potential applications include the ability to correct genetic defects, to treat and prevent the spread of diseases, and to improve crops. However, CRISPR technology still has some serious limitations. One potential complication is that CRISPR editing may lead to other, unintended, genomic changes, known as off-target activity. Moreover, when the CRISPR-Cas system is, intendedly or unintendedly, targeting several different sites in the genome simultaneously, off-target activity can lead to translocations, unusual rearrangement of chromosomes, as well as to other adverse genomic modifications. These, in turn, can have detrimental consequences such as cancer and other pathologies. It is therefore important to detect such potential events in advance. Our goal in this research was to develop a software tool that accurately and reliably quantifies CRISPR-Cas adverse off-target effects, including translocation events, that occur in an editing experiment.
We were motivated by the fact that existing tools are limited as they do not provide statistical evaluation and are therefore not sufficiently sensitive to separate signal from noise in experiments with low editing rates. In experiments utilizing deep-sequencing techniques which have significant levels of background noise, low levels of true off-target activity can get lost under the noise. It is also important to note that most of the current analysis methods are not designed to handle multiplex PCR experiments in which numerous potential targets are analyzed simultaneously. This reality drove the collaboration between the Hendel Lab from Bar-Ilan University and the Yakhini Research Group at IDC Herzliya and the Technion, aimed to develop a measurement approach and related data analysis that are capable of seeing beyond the noise and of detecting adverse translocation events occurring in a genome-editing experiment. The resulting tool, CRISPECTOR, can detect, statistically evaluate, and quantify off-target genome-editing activity.
In our study, our research team assessed 226 off-target sites under diverse experimental conditions to determine the accuracy of the CRISPECTOR tool and demonstrate its advantages. Our novel method is based on a model comparison approach, producing more accurate false-negative rates in sites with weak but significant off-target activity and enabling the analysis of multiplex PCR and NGS data from editing (treatment) vs mock (control) experiments. The first step of the analysis consists of mapping all reads to amplicons. We then consider the deviation from the reference sequence and mark reads as potentially representing edit events. Deviation from the reference can, of course, also be the result of sequencing errors or of other process related issues. In order to discern those reads that actually represent edit events, we use a Bayes classification approach based on data coming from the treatment and mock experiments. When a certain type of deviation from the reference occurs in the treatment with prevalence that is significantly greater than that which is observed in the mock we deem it to be a potential edit event. This is the heart of the machine-learning approach used here, as opposed to simple subtraction which has been used by other tools. User tunable parameters (such as the Bayes priors) can control the extent to which our calling can be considered conservative. With a better understanding of the potential off-target activities of CRISPR genome-editing outcomes we would be able to design alternative editing strategies (for example the use of different CRISPR-Cas systems or guide RNAs) to target the same or similar on-target location with the same required editing effect at the on-target location, while mitigating adverse off-target activities.
One of the exciting features of CRISPECTOR is its ability to detect translocations resulting from CRISPR editing by analyzing NGS data produced by a multiplex PCR using locus-specific primers (such as rhAmpSeq (IDT, Coralville, IA)). CRISPECTOR can analyze DNA reads generated by mixed pairs of primers that amplified the targeted genomic sites as well as adverse translocations that were unintendedly generated during the CRISPR-editing experiment. To do this we slightly modify the read mapping stage, mentioned above, and also identify reads that have primer sequences, on both ends, coming from distinct amplicons. Once putative translocation read configurations are identified, they are scored by CRISPECTOR based on a hypergeometric statistical model and reported to the user, ordered according to the inferred statistical significance. In our study we have experimentally validated the observed translocations and showed independent evidence of their occurrence in human cells. Additionally, we believe that CRISPECTOR can support the analysis of CRISPR genome-editing experiments with multiple guides, which could result in a higher rate of adverse translocations. This makes CRISPECTOR unique in its support for detecting translocation events due to unintended CRISPR-induced genome-editing activity, based on comparative multiplex-PCR data.
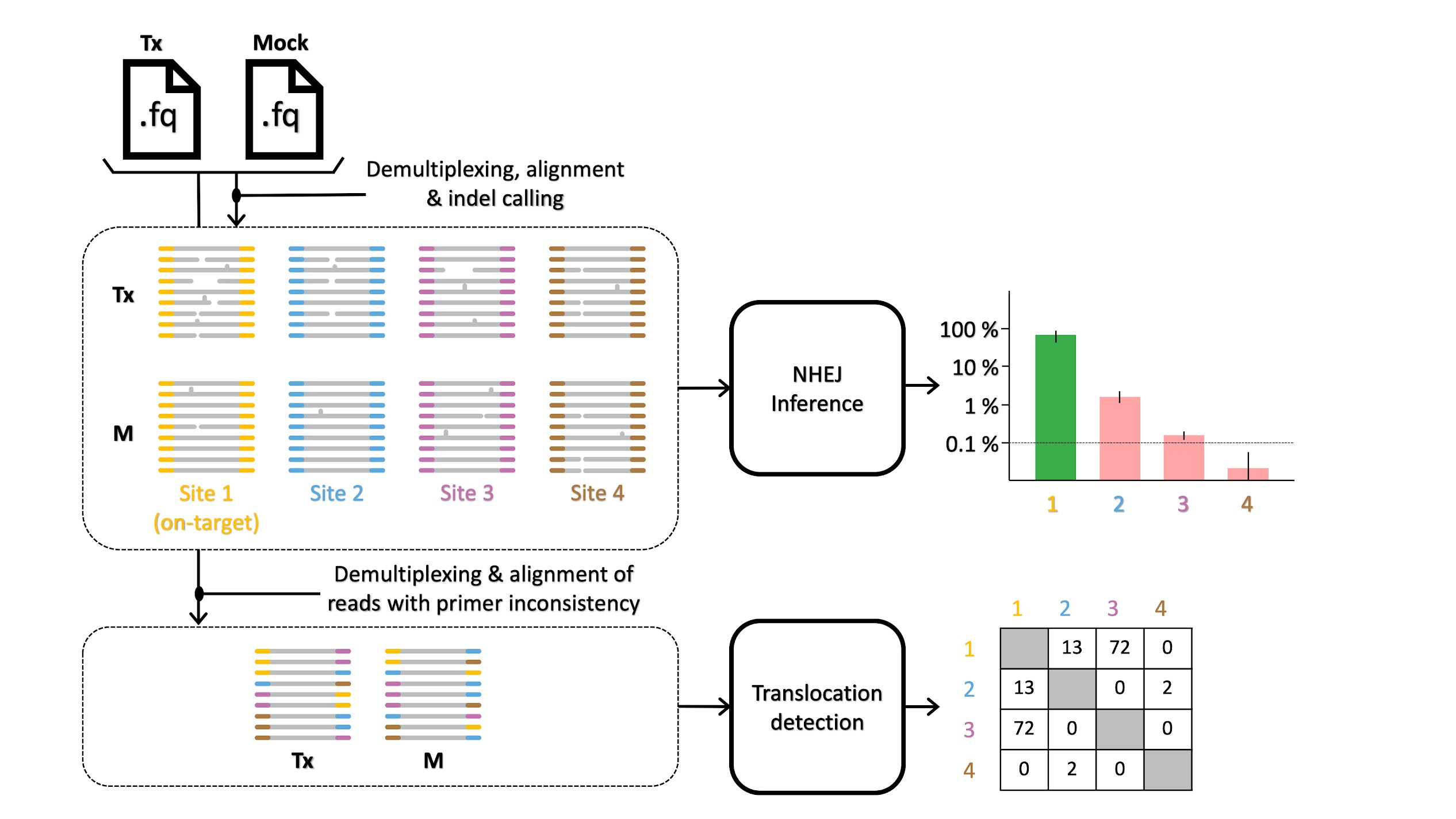
CRISPECTOR workflow: CRISPECTOR assigns each read in the Treatment and Mock (control) FASTQ files to a specific locus of interest or a putative translocation. Then, a Bayesian inference classifier accurately estimates the indel editing activity, and a hypergeometric test is performed to detect translocation reads
In summation, although serious attempts have been made to decrease the amount of CRISPR-Cas off-target activity, much work has yet to be done. We developed a tool that reliably characterizes and quantifies potential CRISPR-induced errors, thus enabling better control and optimization and thereby potentially safer clinical use of genome-editing-based therapeutic approaches. Moving forward, we intend to apply CRISPECTOR to study CRISPR-Cas therapies for genetic disorders of the blood and the immune system, as well as in our cancer immunotherapy studies.
Follow the Topic
Biotechnology
Life Sciences > Biological Sciences > Biotechnology
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
A selection of recent articles that highlight issues relevant to the treatment of neurological and psychiatric disorders in women.
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
This Collection aims to bring together research from various domains related to neurodegenerative conditions, encompassing novel insights into disease pathophysiology, diagnostics, therapeutic developments, and care strategies. We welcome the submission of all papers relevant to advances in neurodegenerative disease.
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in