Critical considerations of sensitivity and specificity for HIV reservoir measurements
Published in Microbiology
Antiretroviral therapy (ART) suppresses HIV viral loads and undetectable viral loads are now understood to mean transmission risk is effectively zero (U=U). However, HIV is a retrovirus that integrates its genome into host cells. Even when viremia is suppressed, HIV proviruses persist in human chromosomes within long-lived CD4+ T cells leading to a persistent and stable reservoir of latently infected cells.
Most latent proviruses are defective due to HIV’s extremely high mutation rate as it enters cells. These sequences leave a record of infection but cannot infect new cells. Roughly 0.1-1% of proviruses are intact and can reinitiate viremia if ART is interrupted. It is crucial to quantify intact HIV proviruses in order to assess progress, whether natural or therapeutic, toward a smaller reservoir and a longer window of remission after stopping ART.
While simple to measure and sensitive, the predominance of defective proviruses means total HIV DNA measurements do not accurately estimate the rebound competent reservoir. The gold standard assay is the quantitative viral outgrowth assay (QVOA) that measures single proviruses that can actually replicate in cell culture. This perfect specificity means sensitivity is more challenging, individuals with small reservoirs can be negative by QVOA but still rebound when ART is stopped.
It is therefore a high priority for the HIV cure field to develop assays which retain the specificity of QVOA but have greater sensitivity to detect replication competent HIV DNA. Based on their rapidity and lower cost, intact proviral DNA assays (IPDA) that define intact HIV sequences based on intactness of two (or more) PCR probes targeted to conserved locations of the HIV genome are now routinely used as endpoints for therapeutic HIV cure trials. These assays achieve a pragmatic balance of sensitivity while approximating the rebound competent reservoir. Although more challenging than IPDA, assays combining probes with near-full length sequencing such as Q4PCR are another emerging technique that reproducibly define intact and defective HIV proviruses and allow interrogation of any discrepancies due to imperfect sensitivity and specificity of each approach.
In this study [Reeves et al. Nat Comm 2023], we compared IPDA and Q4PCR in longitudinal clinical samples from 10 men living with HIV on suppressive ART for 10 years. The detected quantity of proviruses was 40-fold higher using IPDA compared to Q4PCR. IPDA defined viruses had a mean half-life of 108 months, which was not distinguishable from half-lives of total HIV DNA in these participants. On the other hand, Q4PCR defined viruses had a mean half-life of 65 months, with confidence intervals not overlapping with zero, more in line with prior estimates of intact reservoir decay.
In puzzling over the slightly different half-lives, we began considering a subtlety of multi-probe PCR assays: they lack perfect specificity like QVOA. A small percentage of HIV proviruses may be intact at PCR probe locations, but defective elsewhere in the genome. We labeled this crucial third category of HIV proviruses “misclassified defective proviruses” and developed a mathematical model that quantifies the fraction of defective proviruses that would be called intact by a given assay.
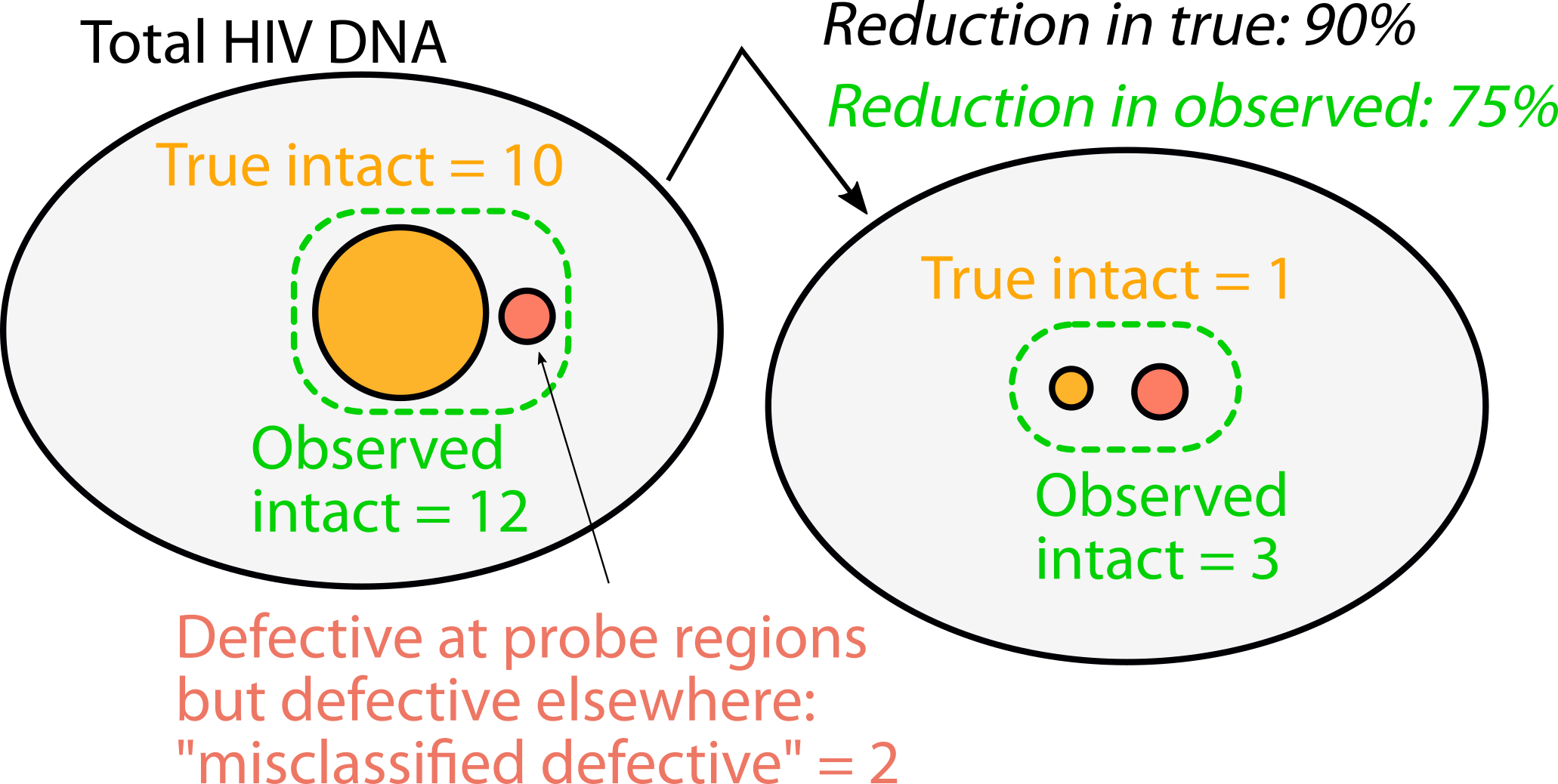
Figure. Schematic depiction of how misclassified defective proviruses could lead to underestimates (simple example: 75% vs 90%) of therapeutic reduction in true intact proviruses.
We then explored the implications of misclassified defective proviruses on reservoir decay measurements. We developed a model which showed that “observed intact sequences” could appear to decay more slowly over time if truly intact sequences continue to decay and slower decaying misclassified defective proviruses increasingly predominate. Decelerating decay of observed intact viruses has indeed been documented in several datasets. We fit our model to intact and defective levels from one data set and estimated an average misclassification fraction of 5% (which generally agrees with estimates derived from libraries of proviral sequences). Therefore, it appeared that misclassification would be one possible explanation of decelerating decay.
Yet, a recently published article [McMyn et al. JCI 2023] demonstrated QVOA+ decay also decelerates. Importantly, our modeling does not disagree with these findings. Because the authors of that manuscript graciously shared their data with us, we also studied a model containing a fraction of very long lived (or even increasing via clonal expansion) intact proviruses and could equally explain observed data. However, we also realized that the presence of a stable long-lived population does not eliminate the impact of misclassification in therapeutic settings.
Most HIV cure strategies including latency reversal agents and immunotherapies attempt to reduce intact proviruses. Our model predicts that reductions in intact proviruses can be underestimated if there are misclassified proviruses that are not also reduced (Figure). We think it is crucial to consider this possibility in clinical trials that appear to demonstrate small intact reservoir reductions. We would not want to miss out on a partially successful therapy that could eventually be improved or combined with others.
A key nuance to the argument can be understood in an analogy to epidemiology. Imagine a diagnostic test for a disease with excellent specificity (>95%). In a population with high prevalence, the small number of false positives will not strongly change the prevalence estimate. However, in a population with low prevalence (for sake of argument can imagine the limit of no prevalence), then most or all tests that return positive are false positives, and the prevalence will be overestimated. Our model mirrors this nuance, in clinical trial simulations, therapeutic reductions are harder to detect in individuals who initially enroll with lower reservoir sizes.
Assays adding additional primer-probe target regions naturally improve specificity but gains in specificity will likely be accompanied by sensitivity loss. We recommend that as next generation multi-probe assays are designed, they should be optimized using proviral libraries to balance misclassification (specificity) and amplification (sensitivity). In the meantime, we also advocate for assays with additional probes and/or full length sequence verification to help estimate changes in reservoir sizes, particularly in clinical trials of promising therapies that admitted minor signals and/or contained participants with relatively small reservoirs initially.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in