Crossmodal Generative AI improves Multimodal Cancer Predictions
Published in Computational Sciences and General & Internal Medicine
Artificial intelligence has shown great promise in cancer diagnostics and prognostics, however, this promise has been difficult to realise in clinical practice due to several reasons related to missingness in multimodal data and lack of reliability and transparency in AI based predictions.
For example, in cancer diagnosis/prognosis, while histopathology slides are routinely collected, transcriptomic assays are expensive, slow, and typically requested only for a small fraction of patients. As a result, many multimodal AI models are trained and evaluated in settings that do not reflect real-world diagnostic workflows. This mismatch raises an important question: can we retain the benefits of multimodal AI when molecular data are unavailable at inference time but available during model training?
In our recent work, we answer this question in the affirmative by investigating whether transcriptomic information can be synthesised directly from routine histopathology images, and whether these synthesised molecular features can meaningfully support downstream clinical prediction tasks. We introduce PathGen, a diffusion-based crossmodal generative model that generates transcriptomic profiles from whole-slide histopathology images, and evaluate its impact on cancer grading and survival prediction across multiple cancer cohorts and types from the Cancer Genomic Atlas.
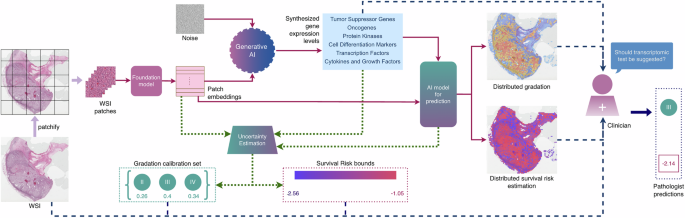
PathGen is designed to learn this crossmodal relationship. Conditioned on patch-level embeddings extracted from whole-slide histopathology images, the model uses a diffusion process to synthesise gene expression features across several biologically meaningful gene groups. To assess clinical relevance, we integrate the synthesised transcriptomic features into a state-of-the-art multimodal prediction framework for cancer grading and survival risk estimation. Across all cohorts, models that combine histopathology images with PathGen-generated transcriptomes significantly outperform image-only models.
Predictive accuracy alone is not sufficient for clinical deployment. To support interpretability, we use co-attention mechanisms that link synthesised transcriptomic features to spatial regions of the histopathology slide. This allows predictions to be mapped back onto tissue morphology, highlighting intra-tumour heterogeneity and providing visual cues that clinicians can interrogate. To quantify reliability, we apply conformal prediction to generate patient-specific uncertainty estimates for both grade and survival risk predictions. Rather than producing single point estimates, the model outputs prediction sets or risk bounds with formal coverage guarantees. This also serves as a measure of predictive fairness across patient demographic subgroups.

Rather than replacing transcriptomic assays, PathGen offers a way to estimate when molecular testing is likely to add value. In future workflows, such models could act as low-cost screening tools, guiding targeted use of expensive molecular assays and supporting more sustainable adoption of multimodal AI. By grounding multimodal learning in routinely available data, and pairing it with interpretability and uncertainty quantification, this work moves closer to AI systems that are not only accurate, but also practical and clinically aligned. By synthesising transcriptomic insight from routine histopathology, PathGen could support earlier, better-informed multidisciplinary team (MDT) discussions by indicating when molecular testing is likely to add clinical value. From a health-economic perspective, this enables more targeted use of costly transcriptomic assays, improving the efficiency and sustainability of precision oncology pathways without replacing clinical judgement.
The code for this work is available open source at GitHub and the data used is available publicly through TCGA and CPTAC portals. An introductory video to multimodal AI is provided below from our team, more such popular science resources are available through our Turing YouTube channel. The research was funded through the Turing-Roche Strategic Partnership.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Cancer in understudied populations Hub
A new Communities’ space to connect, collaborate, and explore research on Cancers, Race and Ethnicity Studies and Mortality and Longevity!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in