Cryo-EM Structure of Lipopolysaccharide Transporter LptDE Opens the Door to Antibiotics Design
Published in Chemistry
The bacterial lipopolysaccharide (LPS) transporter LptDE is not conserved in humans, but plays a pivotal role in many bacteria assisting the essential transport of lipids to the bacterial outer membrane wall. Previous structures of LptDE were all solved by X-ray crystallography.
We report in Nature Communications, 13, 1826 (2022), the first structure done by cryo electron microscopy (Cryo-EM) and the first structure of LptDE from N. gonorrhoeae. Furthermore, it is also the first observation of an LPS transporter “beta-sheet” barrel by Cryo-EM.

The remarkable gymnastics of β-barrel outer membrane proteins
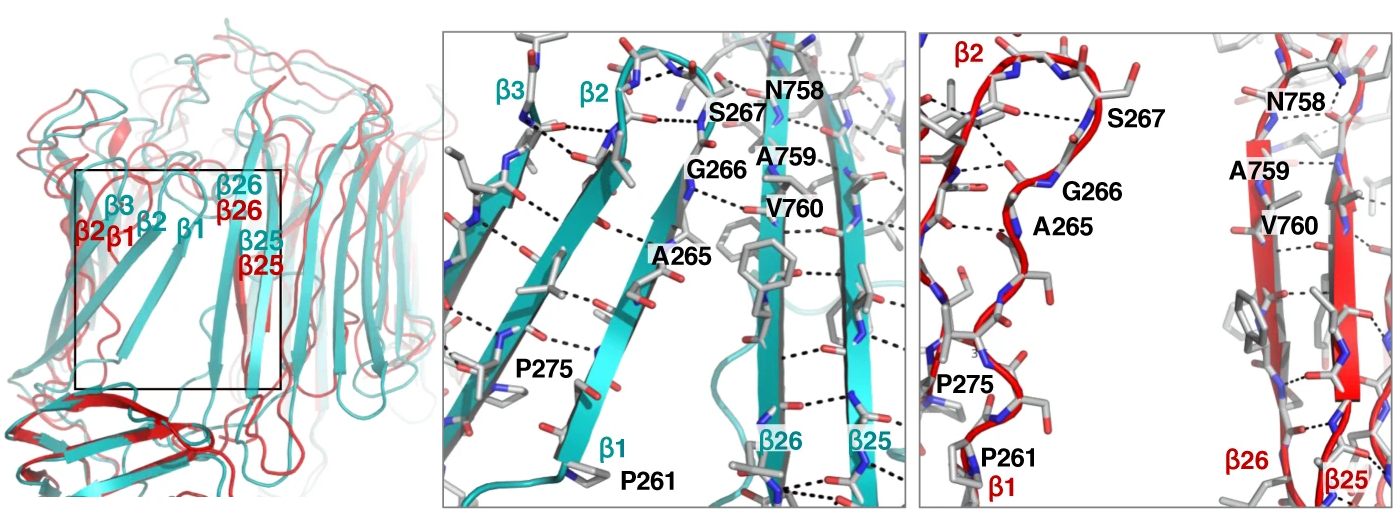
Another exciting feature of the new structure is its experimental confirmation of a theoretically described conformation of the beta barrel (lateral opening), which provides clues at how LPS is released into the bacterial outer membrane (Fig. 2). From this structure, it can be envisioned that the transition to the open conformation allows LPS to be released to the outer membrane.
The design of Pro-Macrobodies (PMbs) for improved cryo-EM and X-rays structures
To reach this high-resolution for Cryo-EM structures, LptDE was stabilized through complexation with computationally designed nanobody-based chaperones (Fig. 1). The technology was developed for this work and called “Pro-Macrobodies”, or PMbs. These chaperones are novel proteins that fuse target-specific nanobodies with a maltose-binding protein via a rigid linker (see our news article here). The linker was designed as two proline residues, giving Pro-Macrobodies their name, and predicted to be rigid via molecular dynamics simulations. The novel tool provides particle enlargement, improves particle alignment and classification, reduces preferred orientation of particles upon grid freezing and ultimately gives higher resolution structures.
The structures of the bacterial transporter LptDE will provide structural basis for the design of novel antibiotics.
References
Botte, M et al. Insight into Lipopolysaccharide Translocation by Cryo-EM structures of a LptDE Transporter in Complex with Pro-Macrobodies. Nature Communications, 13, 1826 (2022).
Bucher, D and Schenck, S. (2021). Pro-Macrobodies for the enhancement of structure research. International PCT Application (No. EP2021/053794).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in