Decoding Salt Tolerance: How Sea Rice HD961 Uses Translation Control to Thrive in Saline Fields
Published in Plant Science
A question born in the salt fields
When I first stood in a saline field watching rows of sea rice HD961 shimmer under the coastal sun, I was struck by a simple question: how does this plant survive where most rice cannot even sprout? Rice feeds over half of humanity, yet salinity quietly undermines its productivity every year. Rising seas and irrigation mismanagement have turned many farmlands hostile to traditional varieties, threatening food security on a global scale. Amid this challenge, HD961—a resilient landrace known as “sea rice”—has become a symbol of hope. It doesn’t merely endure salt; it thrives in it. Understanding how it achieves this resilience could help us design crops that stand strong in an increasingly saline world.
Listening to the language of translation
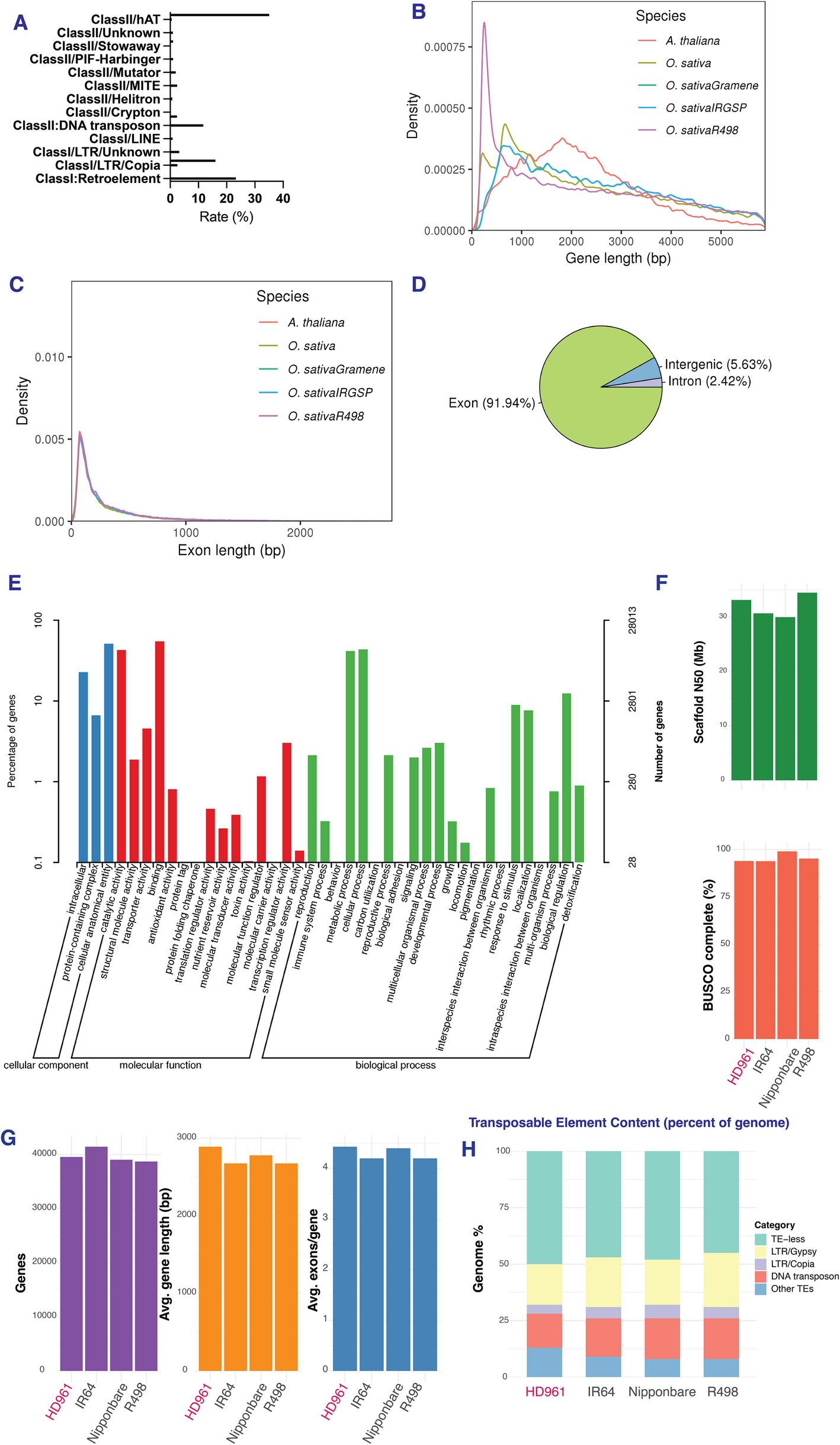
Our team wanted to uncover why, not just at the level of genes but at the level of the decisions a cell makes when it is under stress—what mRNAs to translate, which proteins to prioritize, and how those choices shape survival. We set out to listen to the language of translation itself. The first step was to construct a high-quality chromosome-level genome assembly of HD961 using Nanopore, Illumina, and Hi-C sequencing technologies. The 426.53 Mb assembly achieved exceptional contiguity and captured over 99 percent of conserved eukaryotic genes. When compared with reference genomes such as Nipponbare, R498, and Oryza coarctata, HD961 revealed expansions in gene families associated with ion transport, antioxidant defense, and cell-wall modification—hints that its resilience may be deeply encoded in its DNA.
From the genome to the translatome
But genomes tell only part of the story. What truly fascinated us was how HD961 orchestrates translation under stress. Using ribosome profiling (Ribo-seq) with the precise QEZ-seq protocol, we captured millions of ribosome footprints from control and salt-treated plants. There were moments of frustration: an early analysis mapped reads to the wrong reference genome, producing an unexpected footprint pattern. Only after re-aligning to the HD961 genome did the correct 28–32-nucleotide signature appear—an important reminder that precision matters as much in data as in biology. Once corrected, the patterns spoke clearly.
A strategy of precision, not panic
By integrating RNA-seq and Ribo-seq data, we could see how the plant reallocated its translational machinery under salt stress. HD961 didn’t simply increase overall protein synthesis; instead, it chose where to invest its resources. Antioxidant enzymes such as peroxidases and glutathione-related proteins were translated more efficiently, strengthening the cellular defense system. Ion transporters were tuned finely—some maintained translation, others decreased slightly—reflecting a strategy of balance rather than overreaction. Genes involved in cell-wall biosynthesis, including cellulose synthase A1, showed elevated translational efficiency, likely reinforcing structural stability under osmotic pressure. Enrichment analyses pointed to enhanced secondary metabolism and vesicle trafficking, suggesting that HD961’s response depends more on detoxification and remodeling than on simple ion pumping.
Decoding resilience at the codon level
Digging even deeper, we found that the story extended to the single-codon level. The GCG codon for alanine showed reduced ribosome occupancy at the A site under salt stress, implying faster decoding (Fig. 1). Reporter assays confirmed that sequences rich in GCG codons produced higher translation output when plants were stressed. This subtle shift revealed that HD961 adjusts translation speed at specific codons—tiny edits in timing that together sustain protein synthesis when conditions grow harsh.
eIF2B: a master regulator emerges
One of the most intriguing discoveries centered on the eukaryotic initiation factor 2B (eIF2B), a master regulator of translation initiation. Under salt stress, eIF2B not only showed increased ribosome occupancy but also formed distinct cytoplasmic foci visible in immunofluorescence images. Viability staining verified that these were genuine structures rather than fixation artifacts. We believe these eIF2B-containing bodies may represent liquid-like condensates that gather translation components exactly where they are needed most, maintaining protein synthesis during environmental stress (Fig. 1). Watching them form under the microscope felt like glimpsing the machinery of adaptation itself.
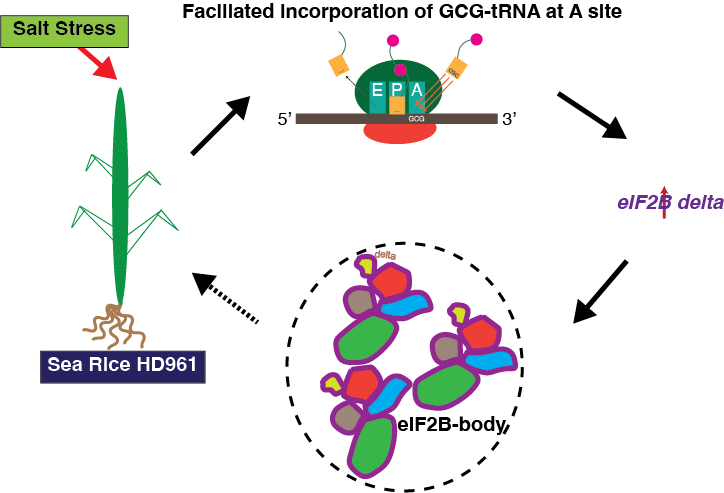
Fig.1 Proposed model of translational adaptation in sea rice HD961 under salt stress. Salt stress promotes efficient incorporation of GCG-tRNA at the ribosomal A site and enhances translation of eIF2Bδ, leading to the formation of eIF2B-containing bodies that help maintain protein synthesis and stress resilience.
From data to discovery
What began as a genome assembly project gradually evolved into a study of how plants think at the molecular level—how they choose, reorganize, and persist. The peer-review process sharpened our analyses, prompting us to refine footprint length distributions, expand translational efficiency data, and make all our resources publicly available. Every correction made the story clearer. All raw data—Nanopore, Hi-C, RNA-seq, and Ribo-seq—are now deposited in NCBI BioProject PRJNA1290000, and the analysis scripts are archived on GitHub and Zenodo to ensure full reproducibility.
Reflections on resilience
Looking back, this project has been a lesson in both patience and perspective. Each layer of discovery—from genome to codon—revealed how finely tuned the language of life can be. Our next goal is to explore whether manipulating eIF2B or codon usage might improve salt tolerance in cultivated rice, translating these molecular insights into tangible benefits for agriculture. For me, HD961 embodies resilience itself: a plant that transforms adversity into adaptation, reminding us that even in salt-stained fields, life finds a way to grow.
Follow the Topic
-
BMC Biology

This is an open access journal publishing outstanding research in all areas of biology, with a publication policy that combines selection for broad interest and importance with a commitment to serving authors well.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biology of neurodegenerative diseases
BMC Biology is calling for submissions to our Collection on the biology of neurodegenerative diseases. This Collection aims to bring together multidisciplinary knowledge to better understand the mechanisms driving progressive neuronal dysfunction and loss.
We welcome studies investigating key pathological mechanisms, including protein misfolding and aggregation, neuroinflammation, oxidative stress, mitochondrial disorders, dysfunction of cellular protein sorting and degradation, altered RNA metabolism, blood-brain barrier impairment, brain vascular dysfunction, contribution of extracellular vesicles and synaptic dysfunction. Research exploring intracellular transport disruptions, neuronal network alterations, and genetic or epigenetic contributions to neurodegeneration is also encouraged.
We are particularly seeking submissions that employ state-of-the-art approaches, such as multi-omics, electrophysiology, high-resolution imaging, (induced) pluripotent stem cell-derived model systems, (e.g. microfluidics 2D co-culture, organoids and assembloids), and animal models to gain deeper mechanistic insights into neurodegenerative processes and identify potential therapeutic targets.
This Collection supports and amplifies research related to SDG 3: Good Health and Well-Being.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Sep 03, 2026
Human microbiome in health and disease
BMC Biology is calling for submissions on our Collection on Human microbiome in health and disease. The human microbiome plays a crucial role in maintaining health. Comprising trillions of microorganisms, including bacteria, viruses, fungi, and archaea, the microbiome influences various physiological processes, such as metabolism, immune function, and even mental health through pathways like the gut-brain axis. Recent advances in sequencing technologies and bioinformatics have enabled researchers to explore the intricate relationships between the microbiome and human health, revealing its potential as a target for therapeutic interventions.
Continuing to advance our understanding of the human microbiome is essential for developing novel strategies to prevent and treat diseases. Significant progress has been made in identifying specific microbial signatures associated with conditions such as obesity, diabetes, and cancer, as well as understanding the impact of antibiotics on microbial diversity. These insights have opened new avenues for personalized medicine, where microbiome profiling could guide treatment decisions and improve patient outcomes.
As research in this field progresses, we can anticipate exciting developments, including the potential for microbiome-based therapies, such as probiotics and fecal microbiota transplantation, to become mainstream treatments. Furthermore, ongoing studies may uncover the role of the microbiome in modulating responses to immunotherapy in cancer patients, leading to more effective and tailored treatment approaches.
Potential topics for submission include, but are not limited to:
The role of the gut, oral, skin, and vaginal microbiome in health and disease
Microbial ecosystems and their impact on the immune system
The gut-brain axis: implications for mental health
The connection between the microbiome and neurodegenerative diseases
Evolution of the human microbiome across different populations
Effects of antibiotics on microbiome diversity
This Collection supports and amplifies research related to SDG 3: Good Health and Well-Being.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in