World AMR Awareness Week: Deciphering the global atlas of antimicrobial-resistant Salmonella using big data
Published in Microbiology and Public Health
Explore the Research

Antimicrobial resistance: a silent pandemic
SPONSOR FEATURES
This study built a global Salmonella genome catalog using 208,233 high-quality genomes. A high-resolution global genetic atlas of AMR in Salmonella was provided, and socioeconomic and environmental factors that can drive the rise of AMR in Salmonella were identified. (Published in Nature Communications on 17 May 2025, https://www.nature.com/articles/s41467-025-59758-3).
The Problem of Antimicrobial Resistance In Salmonella Worldwide
Salmonella enterica (S. enterica) is one of the leading causes of foodborne disease worldwide. In 2019, non-typhoidal Salmonella (NTS), Salmonella Typhi (S. Typhi), and Salmonella Paratyphi caused 215,000, 182,000, and 2,3300 deaths worldwide, respectively, according to the estimates of the Global Burden of Disease Study1. Fluoroquinolone-resistant S. Typhi and NTS and carbapenem-resistant and 3GCs-resistant Enterobacterales were categorized as high-priority and critical-priority pathogens, respectively, in the 2024 World Health Organization Bacterial Priority Pathogens List (WHO BPPL)2. The number of ARGs per isolate increased 1.84 and 2.69 times in both humans and non-human origins in China, as reported in our previous study3. In response to the antimicrobial-resistant Salmonella crisis, we built the Chinese Local Salmonella Genome Database (CLSGDB v2, https://nmdc.cn/clsgdbv2) and released it as an open-access database using ~8,000 genomes4, which has already been accessed 170 thousand times worldwide. Large-scale genomic data and in-depth bioinformatics analysis can greatly expand our understanding of the dynamic trends, evolution, transmission, and drivers of AMR in Salmonella and improve real-time outbreak assessment capabilities5-7. However, the biogeographical patterns of ARGs in antimicrobial-resistant Salmonella remain largely unknown at a global scale. The associations between socioeconomic, climate, environmental, financial sector, infrastructure, health, poverty, trade, and anthropogenic indicators and country-level AMR in global Salmonella have not been revealed.
What did the authors do?
In this study (“A global atlas and drivers of antimicrobial resistance in Salmonella during 1900-2023”, published in Nature Communications on 17 May 2025, https://www.nature.com/articles/s41467-025-59758-3), we aimed to explore the dynamics and evolution of AMR patterns over time in different ecological niches, serovars, sequence types (STs), and identify potential drivers contributing to the spread and persistence of antimicrobial-resistant Salmonella. Moreover, we sought to analyze the associations between different socioeconomic, climate, environmental, health, urban development, and anthropogenic indicators and country-level AMR in Salmonella.
What did the authors find?
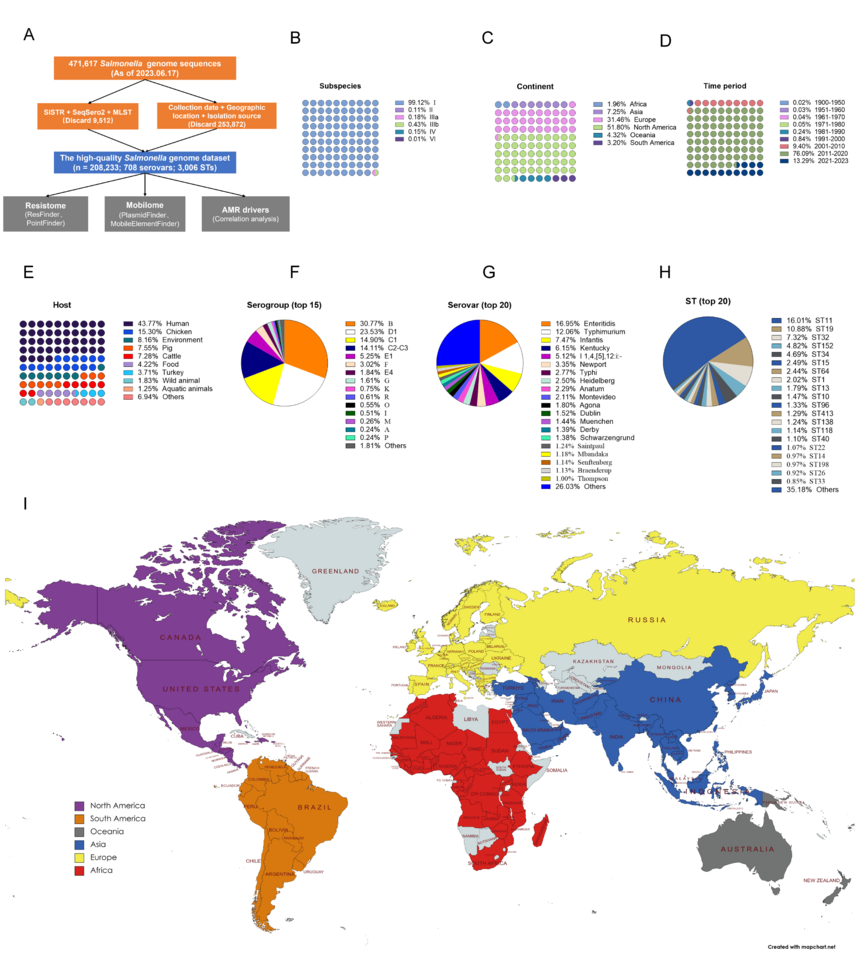
We first built a high-quality Salmonella genome catalog using 208,000 + Salmonella genomes from 148 countries/regions (Figure 1). WGS analysis reveals a complex and worrying global situation. Overall, we found that the geographic distribution of AMR varied depending on the location, source, and serovar. Our results highlight vital spatial disparities in AMR in Salmonella and provide estimates for countries/regions lacking genomic surveillance data.
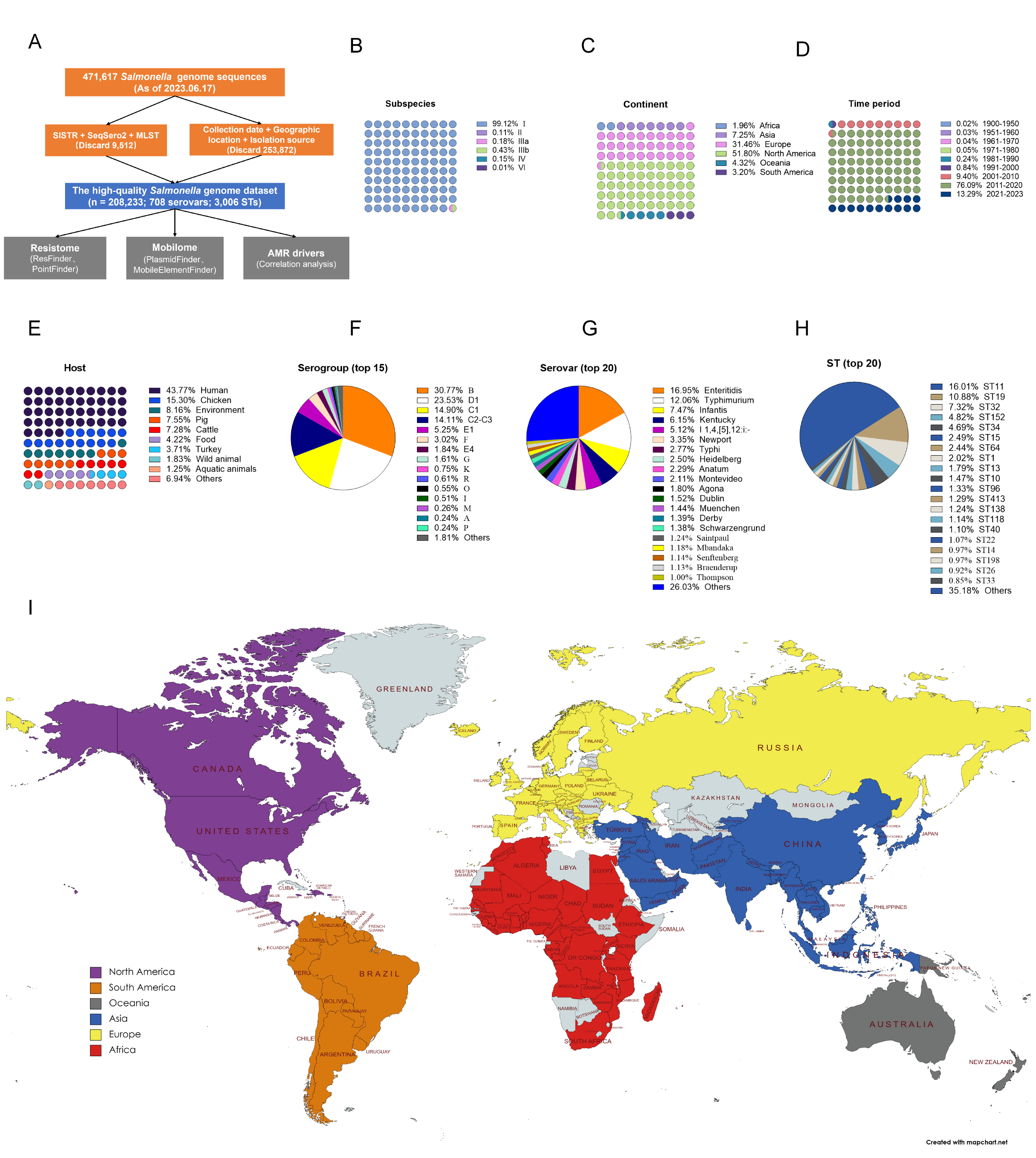
Figure 1: Harnessing a global high-quality Salmonella genome catalog to combat antimicrobial resistance and inform outbreak responses.
Regional hotspots of fluoroquinolone non-susceptible were predicted in India, China, the USA, Brazil, Nigeria, Chile, and South Africa. The prevalence of fosfomycin resistance was higher in South America (16.26%), Asia (10.47%), and North America (9.09%) than in other continents (R% < 5%). Regional hotspots (R% > 10%) of fosfomycin resistance were predicted in Brazil, China, Canada, Chile, and Germany. The prevalence of fosfomycin resistance rates was higher in pigs, turkeys, chickens, and food than in humans and the global average. MDR rates vary across continents; the highest MDR rate was found in Asia (50.96%), followed by North America, South America, and Africa, all of them above the global average (34.05%). The prevalence of resistance rates for individual antimicrobial classes differed among different sources. Host hotspots (R% > 40%) of MDR were predicted in turkeys, chickens, and pigs. High fluoroquinolone non-susceptible rates were observed in both animals, food, and the environment. The prevalence of resistance to polymyxin was above 0.65% (global average) in turkeys (5.87%), wild animals (1.29%), and pigs (1.21%). The prevalence of resistance to macrolides was above 1.11% (global average) in pigs (2.22%), aquatic animals (1.76%), chickens (1.35%), and wild animals (1.13%).
At the global level, the trend is unmistakably upward of diverse wave shapes. The most alarming rise is in resistance to fluoroquinolones, a class of antibiotics critical for treating severe salmonellosis. This increase is particularly sharp in Asia, Oceania, and South America. When we zoomed in on different sources, a worrying pattern emerged: the resistance levels and the average number of ARGs per genome are skyrocketing in chickens, food, wild animals, and the environment. Serovars like Agona, Dublin, I 1,4,[5],12:i:-, Muenchen, Senftenberg, and Mbandaka are at the forefront of this dangerous wave.
Globally, the acquired ARGs increased from 1.16 per genome in 1900-1950 to 2.87 per genome after 2020. From the genomic analysis, the number of ARGs per genome increased sharply with increasing years observed in chickens, food, wild animals, and the environment. Mobile genetic elements analysis also indicated an increasing trend in the number of plasmid replicons over time in some serovars, for example, S. Kentucky, S. Thompson, S. Mbandaka, S. Montevideo, S. Muenchen, S. Sainpaul, S. Heidelberg, and S. Dublin.
In addition, we also used publicly available S. enterica genomic data and 115 indicators for correlation analysis, and identified that agriculture & rural development, climate change, the environment, urban development, infrastructure, socioeconomic factors, and antibiotic consumption in both humans and animals contribute to the evolution of AMR in Salmonella.
What are the limitations of this study?
Despite providing valuable insights, this study relied on publicly available genomes from NCBI and our Chinese Local Salmonella Genome Database (CLSGDB v2, https://nmdc.cn/clsgdbv2), which may not reflect actual AMR levels in different sources within countries/regions. In addition, there is latency in the genomic data we use. Future studies should consider comparing the evolutionary trajectory of AMR in Salmonella due to the influence of a single variable.
Conclusions
This study presents the valuable to-date genetic atlas of antimicrobial-resistant Salmonella in the concept of “One Health”, including humans, animals, food, and the environment across the world. To our knowledge, our study provides a valuable big data catalog and systematic study reporting the temporal and spatial dynamics of AMR in S. enterica from diverse sources in 148 countries/regions across six continents, and fills in the data gap on the spatiotemporal characteristics of resistance across different antimicrobial classes at the global scale. This work enhances our understanding of the dynamic trends, evolution, transmission, and drivers of AMR in Salmonella over the past 100 years. These data provide valuable information for understanding the transmission dynamics and evolutionary trajectories of Salmonella, and public health decision-makers prioritizing interventions for foodborne diseases and preventing the spread of antimicrobial-resistant Salmonella.
Reference
1 GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 400, 2221-2248, doi:10.1016/S0140-6736(22)02185-7 (2022).
2 World Health Organization. WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. World Health Organization, doi:https://www.who.int/publications/i/item/9789240093461 (2024).
3 Wang, Y.N., Liu, Y., Lyu, N., et al. The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl Sci Rev 10, nwac269, doi:10.1093/nsr/nwac269 (2023).
4 Wang, Y.N., Xu, X.B., Zhu, B.L., et al. Genomic analysis of almost 8,000 Salmonella genomes reveals drivers and landscape of antimicrobial resistance in China. Microbiol Spectr 11, e0208023, doi:10.1128/spectrum.02080-23 (2023).
5 Wang, Y.N., Xu, X.B., Zhu, B.L., et al. Enhanced Genomic Surveillance Is Essential for Effective Salmonella Outbreak Response. China CDC Weekly 7, 880-881, doi:10.46234/ccdcw2025.151 (2025).
6 Wang, Y.N., Liu, Y., Zhu, B.L., et al. Salmonella enterica ST8333 Was Isolated as early as July 2015. China CDC Weekly 7, 1347-1349 (2025).
7 Lv, P.P., Pei, Y.H., Jiang, Y., et al. Genomic insights into antibiotic-resistant non-typhoidal Salmonella isolates from outpatients in Minhang District in Shanghai. Commun Med 5, 228, doi:10.1038/s43856-025-00950-3 (2025).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in