Decoding the genomes of coral reef symbionts
Published in Ecology & Evolution

Our paper published in Communications Biology can be found here.
“Do you go diving at the Great Barrier Reef regularly to collect your samples?”
This is the question that I always get asked when I tell people that I am working on the genomes of algae that grow symbiotically with corals. My response of “nope, never” appears to stun many, and in some cases, kills the conversation instantly. Welcome to my world of genomics and computational biology.
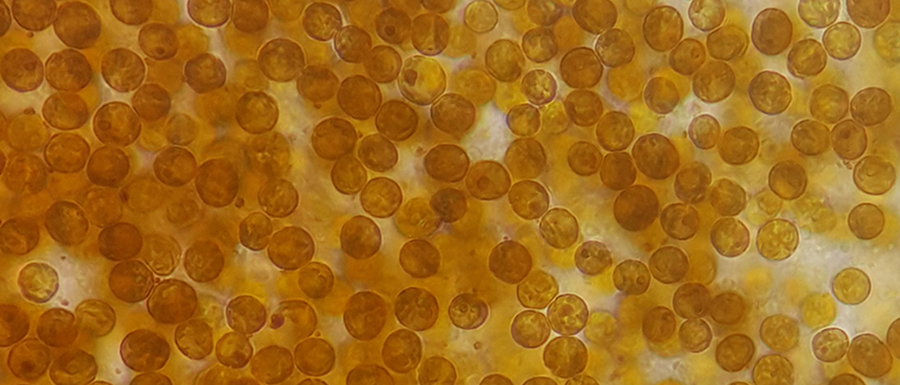
During my postdoctoral stint at Rutgers, I learned that genomes of dinoflagellates are some of the most peculiar known, so peculiar that some researchers had called dinoflagellates aliens from outer space. These genome features include non-canonical nucleotides, unsual intron-exon splice signals, abundant repetitive elements, extensive RNA editing, and the list goes on. Perhaps more intriguingly, these microscopic algae/phytoplanktons/grazers/mixotrophs/photoautotrophs or whatever-you-call-them could have a large genome that is more than 70 times larger than a human genome. Another blog here also highlighted the peculiarity of diverse dinoflagellates. To me, dinoflagellate genomics is fascinating, challenging, and one of the final frontiers in eukaryote genomics.

When Mark Ragan in 2013 advertised the Bioinformatics Fellowship position funded by the Great Barrier Reef Foundation to work on the genomes of the symbiotic dinoflagellates (Symbiodinium) at the University of Queensland’s Institute for Molecular Bioscience, I applied for the position with no hesitation. The position is part of the Sea-quence Project of the Reef Future Genomics (ReFuGe) 2020 Consortium, which aimed to generate core genetic data from multiple species of corals, and the associated Symbiodinium and microbes. It is well known that Symbiodinium are critical symbionts to corals without which corals would starve and eventually die (i.e. coral bleaching). This motivated me even more to get involved in this project.
As with any large research consortium, it took us a little while to decide on which coral genomes to sequence, as one would campaign for their favourite species. It was easier for us to determine which Symbiodinium isolates to focus on: (a) Symbiodinium goreaui of type C1 and (b) Symbiodinium kawagutii of clade F. Type C1 is the most dominant Symbiodinium type in the Great Barrier Reef (and in Indo-Pacific and Caribbeans), and both isolates were readily available to the Consortium. The “why we chose these isolates” question is less critical when no Symbiodinium genome data were available at that stage. We simply wanted to generate high-quality reference genome data for Symbiodinium.
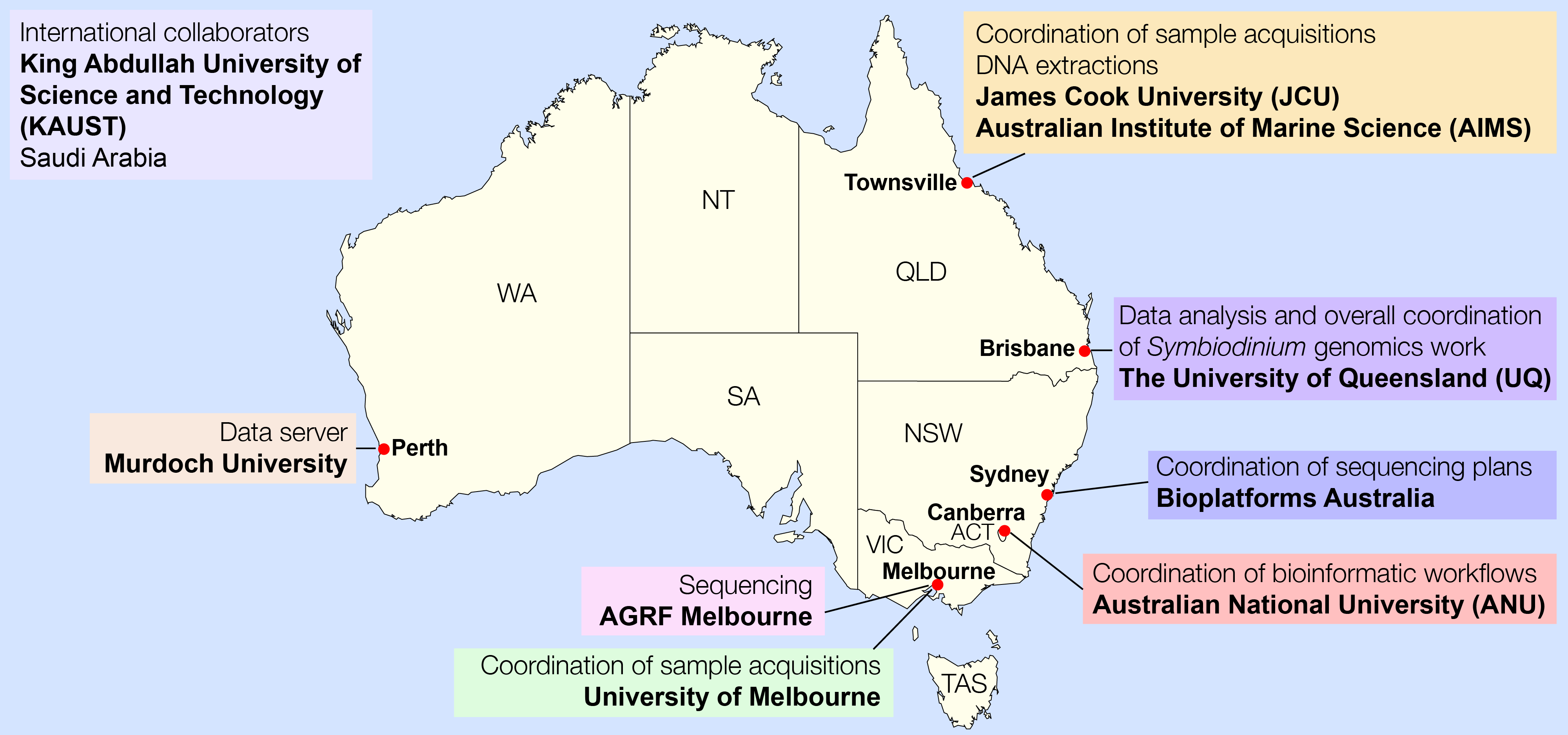
The genomic DNA from these Symbiodinium isolates was then extracted at James Cook University and the Australian Institute of Marine Science in Townsville (Queensland). We coordinated the sequencing strategy with Bioplatforms Australia in Sydney (New South Wales), the sequencing was carried out in Melbourne (Victoria), the sequence data were ported via a data server in Perth (Western Australia), and core data analysis was done in Brisbane (Queensland). We received our very first batch of HiSeq 2500 sequence data on 25 September 2013. It sounds like a lot of effort, and indeed it was.
Given the complexity of dinoflagellate genomes, we spent a long time to carefully optimise the genome assemblies, and to design a customised, dinoflagellate-specific workflow for genome annotation. As time went on, we needed more hands on deck. Through a funded and related project from the Australian Research Council, postdoc Huanle Liu together with two PhD students, Timothy Stephens and Raúl González-Pech, came on board. As a team, we drove this project from data analysis, design and optimisation of analytic workflows, debugging computer codes, decoding cryptic error messages, arguing about who used up all the disk space on the supercomputer, to consoling each other when weeks of long-running computation was prematurely terminated with no way to resume it. Throughout this project, we have used up easily more than two million CPU hours.
As the time went on, Symbiodinium genomes started to become available from other research groups, and these data became our test datasets. We had to make sure that the S. kawagutii genome published in November 2015 was indeed from the same isolate we have used, and we later incorporated all these data together to refine our assembly. Huanle and I went down to visit Sylvain Forêt and Emily Hua Ying at the Australian National University in Canberra for two days in November 2015 to brainstorm and streamline our annotation strategies with their workflows designed for coral genomes. The weather in November, leading to the austral summer, can be quite hot in Australia. I had a heat stroke on a 36°C-day and puked on the street (sorry, Canberra!), but it was well worth it.
Once we were happy with the assembled genomes and predicted gene models, the comparative analysis using other Symbiodinium genomes to probe positive selection in gene functions and other analyses took about six months. By the time we finally put everything together in the form of a manuscript draft and started circulating to the other co-authors it was 3 July 2017, a little less than four years since we received the first batch of sequence data. We had a draft ready for submission two months later, and put it as a bioRxiv preprint to avoid further delay in data release. This paper was first submitted to Communications Biology in December 2017, and underwent a rigorous, three-round review process. We are grateful to the three anonymous reviewers who provided constructive feedback that have helped us to improve the manuscript. We hope that our contribution here will help guide researchers to investigate the resilience of coral-dinoflagellate symbiosis (thus of the coral reefs), and more broadly the biology and evolution of dinoflagellates.

The journey for us to get here has been long, arduous, and it involved many sleepless nights. We had a lot of fun too. Through the ReFuGe Consortium, I got to attend meetings at some of the most amazing places. Some highlights include our meeting in Saudi Arabia, and snorkeling around the pristine reefs in the Red Sea.

I finally got to see some of the magnificent reefs around the Great Barrier Reef in Orpheus Island, but not without first doing a ridiculous photo pose with a life jacket and a noodle.

Our final meeting was in November 2016, at James Cook University’s Marine Research Station at Orpheus Island. We even did a funny group shot after we learned about the change of leadership in the US that day.

Sadly, Sylvain passed away in December 2016. We dedicate this paper to Sylvain, who is sorely missed.
All data generated from this paper and from the ReFuGe Consortium are available at http://refuge2020.reefgenomics.org/.
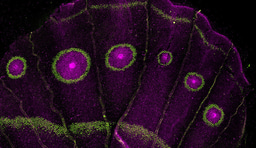
Poster image photo: Symbiodinium goreaui cells inside the coral polyps of Acropora tenuis by Katarina Damjanovic.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in