Defining cell surface receptors recognized by conjugative plasmids using high-throughput assays
Published in Microbiology

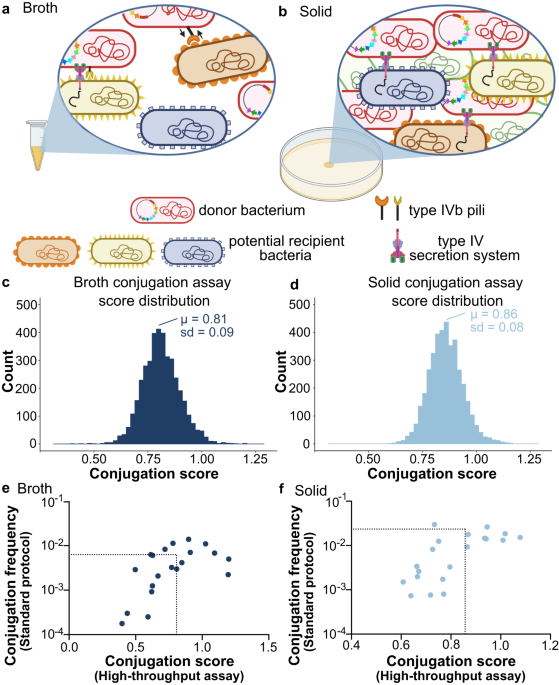
Conjugation, a key process in horizontal gene transfer, plays an important role in bacterial evolution and the emergence of antibiotic resistance. To achieve conjugative transfer, it is essential for donor and recipient cells to come in close proximity, leading to mating pair formation (MPF). Alongside MPF, a generally overlooked step is mating pair stabilization (MPS). This MPS is particularly crucial in unstable environments to maintain the interaction between donor and recipient bacteria, enabling the effective transfer of DNA. The MPS process depends on the presence of adhesins on the donor bacterium’s surface or structures such as pili, which engage with specific components on the outer membrane of recipient bacteria.
Conjugative plasmid TP114 is noteworthy for its high transfer efficiency within the mouse gut microbiota. The conjugative transfer of TP114 whether in vivo or in broth relies on an accessory type IVb pilus (T4Pb) that plays a role in MPS while it is dispensable for conjugation on a solid media. A distinctive characteristic of the T4Pb encoded by I-complex conjugative plasmids is the presence of a multiple DNA inversion system known as a shufflon. This system extends from the distal part of the pilV gene to the rci gene, which encodes a site-specific recombinase also referred as a shufflase. The pilV gene is followed by several DNA segments bordered by rci-recombination motifs. This arrangement allows for either individual or collective reordering, modifying the 3'-end of pilV. Depending on the T4Pb, it results in the creation of 2 to 8 distinct PilV variants, of which one is believed to be displayed at the tip of the pilus.
Although the functions of genes encoded by conjugative plasmids have been thoroughly investigated, the contribution of genes within the recipient chromosome is still not well understood. Addressing this gap, our research explored the genetic prerequisites in recipient cells necessary for the conjugation of IncI2 plasmid TP114. We developed a novel high-throughput methodology to performed transfer assays with approximately 4,000 single-gene deletion mutants of Escherichia coli. Interestingly, when conjugation occurred on solid media, we found that genes in recipient cells that reduced transfer rates did not correlate with any particular cellular functions. However, in broth-based assays were significantly dependent on the lipopolysaccharide biosynthesis pathway. This finding implies that the various PilV forms recognize distinct elements in the recipient cells lipopolysaccharide, facilitating conjugation in unstable environments where factors like movement, fluid dynamics, and external factors could otherwise impede bacterial interactions.
Our study further uncovered that each of the eight PilV variants of TP114 can facilitate its transfer in at least one of the five core-oligosaccharide prototypes found in E. coli, namely, types R1, R2, R3, R4, and K-12. Moreover, by examining the K-12 LPS core-oligosaccharide region through the use of selected deletion mutants or the introduction of exogenous waa genes from other core types, we were able to pinpoint specific disaccharide structures that are recognized by six out of the eight PilV variants of TP114.
Considering that PilV adhesins receptor structures are shared across various bacterial genera, such as Salmonella, Shigella, Klebsiella, Citrobacter, and Vibrio, some of which include pathogens implicated in healthcare-associated infections, the eight PilV variants within TP114's shufflon might broaden its ability to transfer between hosts. IncI2 plasmids like TP114 are known for spreading among Enterobacterales including E. coli, Salmonella, and Klebsiella, and can even transfer to certain Pseudomonadales. Leveraging this expanded host range could be pivotal in developing or improving new methods to combat antibiotic resistance, using conjugation-mediated effector delivery by live biotherapeutics. Additionally, exploring inhibitors of conjugation or MPS could be crucial in curtailing the spread of multidrug resistance. Our comprehensive, high-throughput method is a powerful tool for investigating other mobile genetic elements, providing insights into critical host cell factors in their dissemination.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in