Studying bacterial conjugation reveals a proficient DNA delivery machinery for microbiome editing
Published in Microbiology
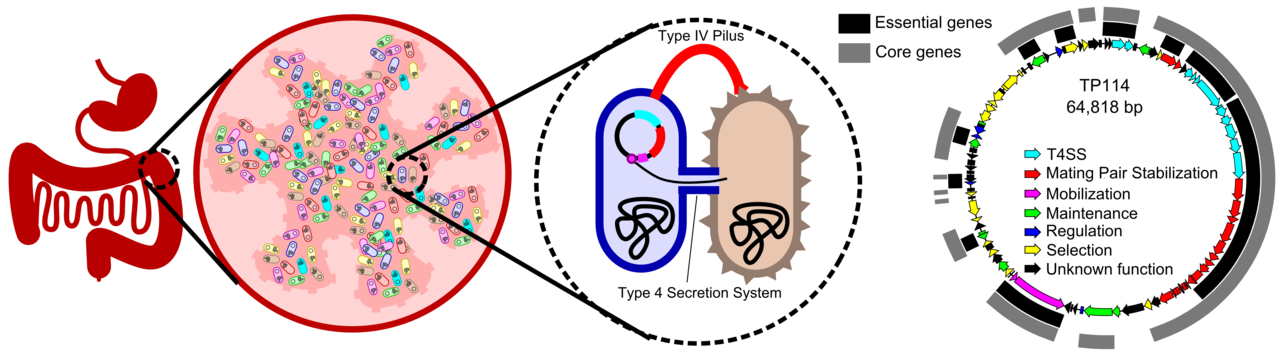
The intestinal microbiota, with its diverse and dense populations of bacteria, is thought to be a prolific environment for horizontal gene transfer, especially through bacterial conjugation. This could have important consequences on the dissemination of antibiotics resistance genes to opportunistic pathogens. But what do we really know about the mobility of genes in the gut microbiota? Are all conjugative plasmids equally capable of disseminating in this environment?
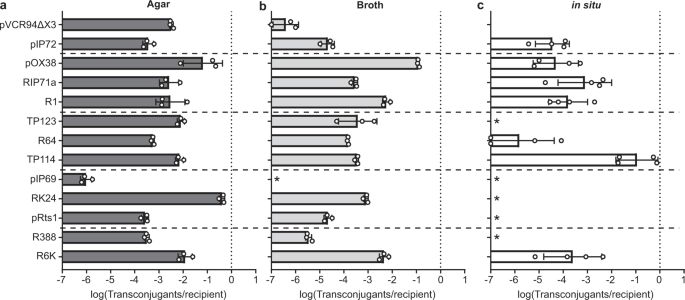
In our recent study, we quantified transfer rates of many conjugative plasmids isolated from a variety of incompatibility groups present in Enterobacteriaceae. To our surprise, while all these plasmids were active in vitro, most of them failed at transferring in the mouse intestinal microbiota; including the highly efficient laboratory workhorse RP4. Among the few plasmids that yielded transconjugants, only IncI2 plasmid TP114 reached high conjugation rates.
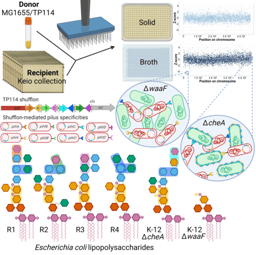
We next investigated the genetic features of the TP114 plasmid required for conjugation using a high-density transposon mutagenesis library. We performed conjugation assays in vitro, on agar, or in vivo, in the mouse intestinal microbiota, to identify genes that are essential for TP114 transfer in each condition. Interestingly, genes involved in the assembly of a specialized extracellular structure called the type IV pilus (T4P) were found to be essential for conjugation in the gut but not on a solid support. Since the T4P is involved in mating pair stabilization (MPS) in I-complex plasmids, we thus wondered if this mechanism could also be required for other plasmids to transfer in the gastrointestinal tract. Using bioinformatics tools and in vitro experiments, our results suggest that all plasmids that transfer in the gut microbiota use MPS, while plasmids that failed to do so were missing key genes for this process.
Our work provides important insights into the mechanisms involved in conjugation in the gut. This knowledge could be useful to monitor the likelihood of antibiotic resistance gene dissemination by specific plasmid families. Also, the identification of TP114 as proficient conjugative machinery paves the way for a next generation of drugs based on bacteria performing microbiome editing by delivering genetic payloads to microbial populations of the gastrointestinal tract. Stay tuned!
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in