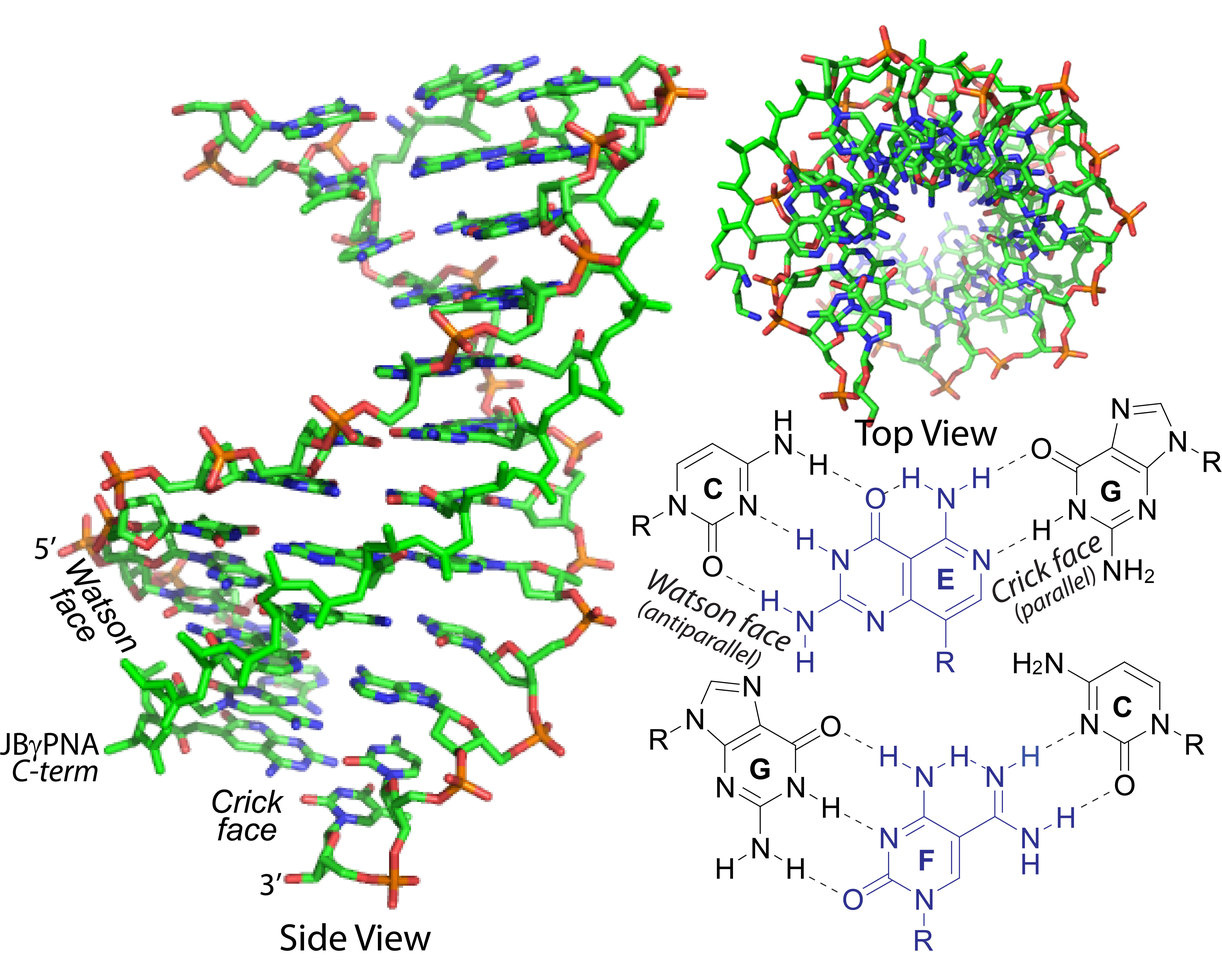
The Watson-Crick base-pairing (hydrogen-bonding) interactions, A with T or U and C with G, are synonymous with nucleic acid recognition. Employed by nature in the storage and transmission of genetic information and by chemists and biologists in the recognition of genetic materials, they are rarely applied in the recognition of double helical DNA or RNA. One reason is that most oligonucleotide molecules developed to date do not have adequate binding free energy to invade DNA or RNA double helix—a necessary requirement for establishing directional hydrogen-bonding interactions with the designated nucleobase targets. Peptide nucleic acid (PNA), a nucleic acid mimic developed by Nielsen and coworkers in the early 1990’s that is comprised of an achiral polyamide backbone, holds considerable promise in this regard because of its tight binding and charge-neutral backbone. However, even with the latest advance, in the development of a conformationally-preorganized backbone, achieving sequence-unrestricted recognition of double helical DNA or RNA under physiological conditions remains a distant goal.
To address this challenge, we developed a new set of bivalent recognition elements that are capable of forming directional hydrogen-bonding interactions with nucleobases in both strands of the DNA or RNA double helix and appended them to a conformationally-preorganized gammaPNA backbone. These so called “Janus bases” differ from that others that have been developed to date in that they are uniform in size and shape and have the appropriate chemical functionalities and tautomeric structures for optimal binding with the canonical as well as the noncanonical base-pairs (16 in total). To test the feasibility of this design concept, we subjected it to the most stringent test in the invasion of DNA and RNA containing C-G and G-C base-pairs. Our data showed that not only are they able to invade such a thermodynamically stable target, but to do so under a physiologically relevant condition. Such a design is possible because of the asymmetry in the right-handed helically-induced gammaPNA backbone, which prevents the self-complementary Janus bases from hybridizing to each other.
With this success, the next step will be to determine whether the other Janus bases could be synthesized and programmed to bind to their designated base-pair targets in DNA or RNA double helix. There is optimism that such a goal could be accomplished since all 16 Janus bases were designed in the same way. With an increase in the number of hydrogen-bonding and in the degree of base-stacking interactions, along with backbone preorganization, there is a likely chance that significantly shorter probes, in the range of 3-6 nucleotides, in the millamolecular range, could be developed for the manipulation of RNA structure and function.
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
f-block chemistry
Publishing Model: Open Access
Deadline: Feb 28, 2026
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in