Discovering learned interactions in proteins
Published in Physics and Computational Sciences
Proteins are the molecular machines of life: they fold, move, and interact in ways that determine the function of entire cells. Understanding these motions in microscopic detail is key to explaining fundamental biological processes and designing new drugs and materials. For decades, researchers have worked to develop simulation methods capable of capturing these intricate dynamics with high accuracy. Yet, simulating the motions of large biomolecular systems remains immensely demanding: A single simulation can require months or years of computation on supercomputers.
The advent of neural network potentials (NNPs) brought a breakthrough promise: achieving quantum-level accuracy at molecular dynamics speed, bridging the gap between the physical world and computational modeling. Accelerating molecular dynamics even further, reduced resolution models with fewer degrees of freedom, known as coarse-grained models, achieved unprecedented performance thanks to the expressivity of NNPs.
Yet, using NNPs comes at the price of interpretability. Unlike traditional models, which express their physical assumptions through transparent functional forms, NNPs tend to obscure their inner workings behind thousands of learned, and often opaque, parameters. Our motivation for this work stemmed from a simple but persistent question: Can we look inside this black box and understand what the network actually learns about physical interactions in proteins?
From mystery to many-body interactions
In physics-based models, every term carries a meaning: Two-body potentials describe pairwise interactions, three-body terms encode angles, and so forth. In contrast, neural networks learn an abstract representation of the molecular system. The features captured in these representations do not necessarily align with the physically defined interactions we know from theory. Still, we believed that traces of this underlying physical structure might remain hidden within the network. If only we knew how to extract them.
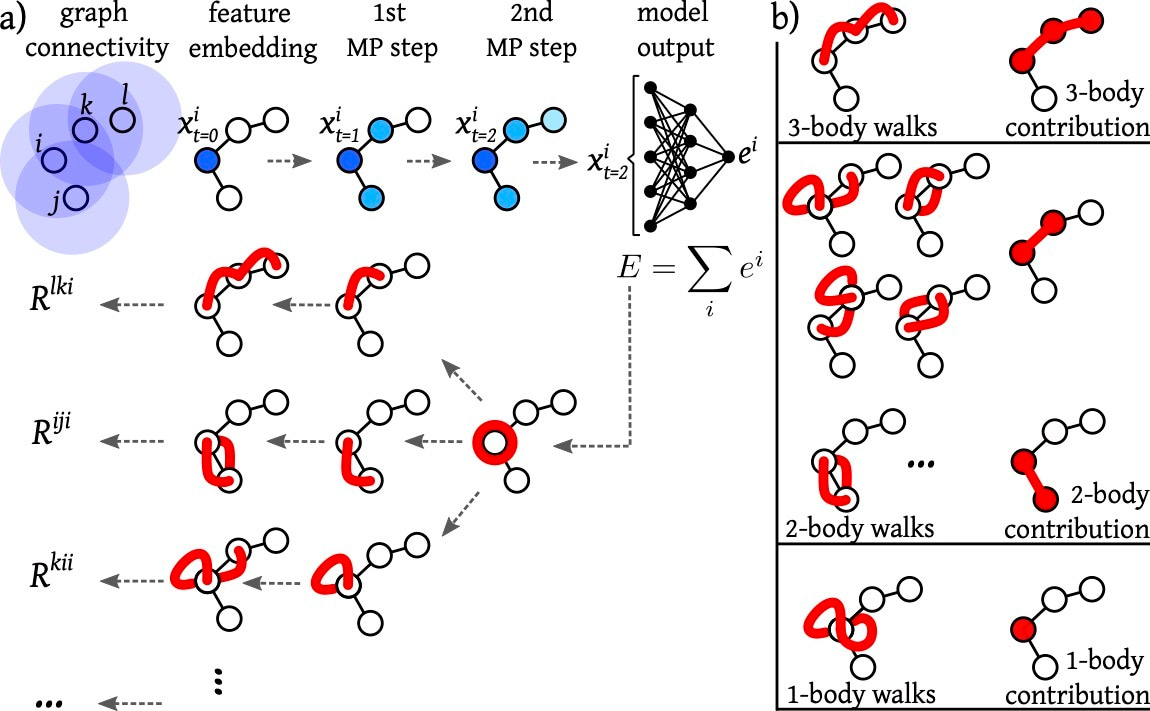
To peer inside the neural network, we built upon the explanation framework Layer-wise Relevance Propagation for graph neural networks. Since molecular structures can naturally be represented as graphs, this approach was particularly well-suited to our problem. We developed a post-processing procedure that transformed the model’s relevance maps into a human-readable form based on the coarse-grained representation. This required linking the abstract representations learned by the NNP to the physically interpretable framework of many-body interactions.
Using this method, we analyzed NNPs trained on different molecular systems. Now we could quantify how strongly each model relied on different types of interactions based on the two-body, three-body, or higher-order terms provided by the explanation method. What emerged was a surprisingly structured picture: Although the networks were never explicitly told about these interactions, they re-discovered them, assigning varying importance to different many-body contributions depending on the system and training regime. This allowed us to “open” the network and visualize, for the first time, the learned many-body interactions of NNPs in protein folding processes and mutation.
A step toward interpretable protein folding
The implications go beyond curiosity. As machine learning becomes an integral part of materials modeling and molecular simulation, trust and interpretability become essential. By learning how NNPs internally balance physical interactions, we can better design them, debug them, and understand when they might fail.
This work represents one small step toward making NNPs explainable, bridging the gap between the language of physics and the language of neural networks. This opens the door to getting physical insight into unknown systems from neural network predictions. For instance, our methodology may help to better understand processes like protein folding and identifying the crucial interactions driving those processes.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Ask the Editor – Space Physics, Quantum Physics, Atomic, Molecular and Chemical Physics
Got a question for the editor about Space Physics, Quantum Physics, Atomic, Molecular and Chemical Physics? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in