Domain reversal: A type VII-secreted lipase toxin with reverse domain arrangement
Published in Microbiology and Cell & Molecular Biology
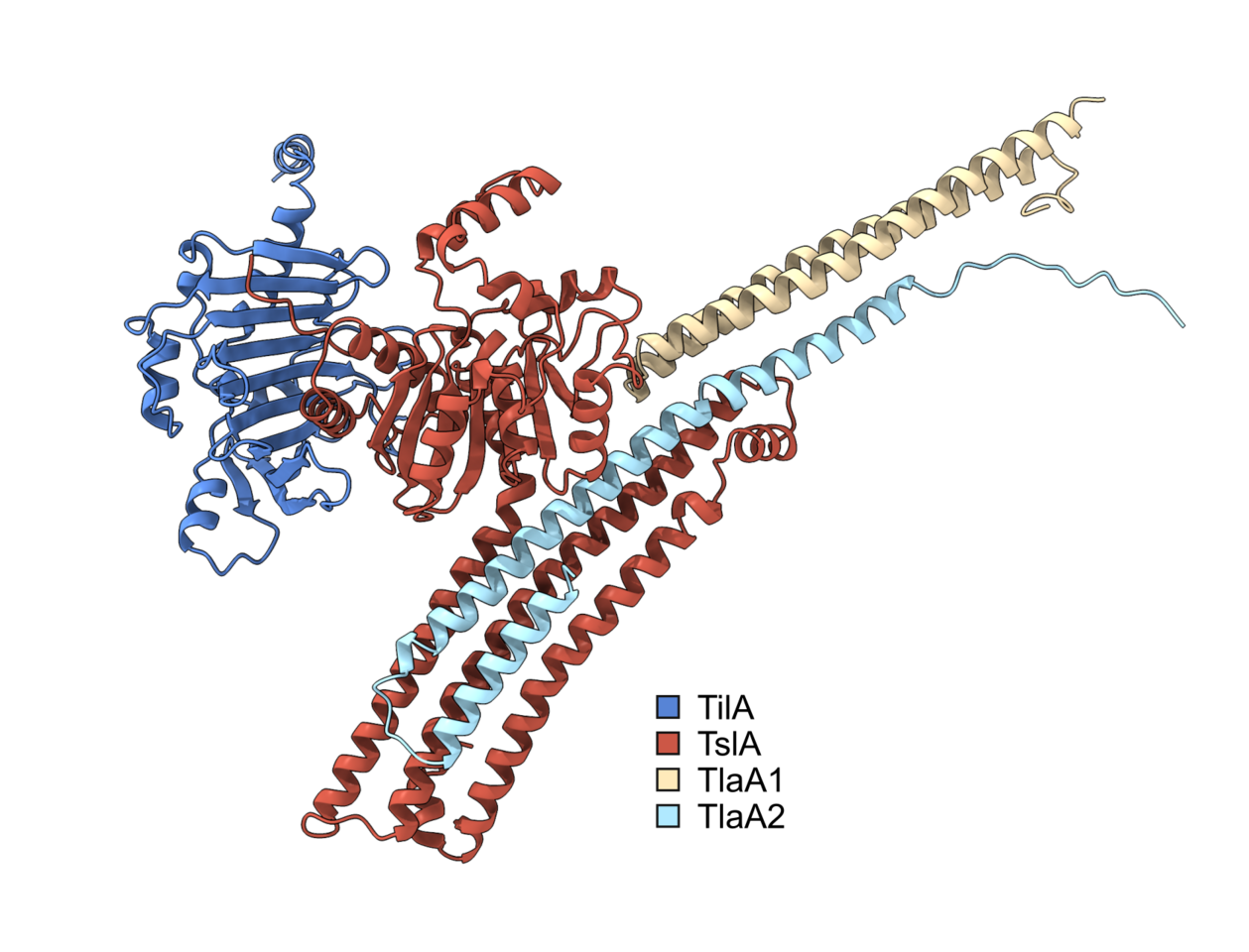
The type VII secretion system (T7SS) is a mechanism by which proteins are exported out of the Gram-positive cell, and it is found in several prominent pathogens such as Mycobacterium tuberculosis and Staphylococcus aureus. In S. aureus, the Palmer lab has shown that large proteins secreted by the T7SS can be used for interbacterial competition, mediated by C-terminal toxin domains present on the substrates. To identify novel substrates of the S. aureus T7SS, the secreted proteome of a wild type (WT) strain was compared to that of a T7SS mutant. Several proteins were more abundant in the supernatant of the WT, suggesting they may be secreted by the T7SS. However further analysis revealed that most of these were not T7SS substrates, and their absence in the supernatant was ascribed to pleiotropic effects associated with loss of T7SS activity. One protein, however, could not be ruled out as a substrate, despite sharing no sequence features with known T7SS substrate proteins1. The aim of my PhD was to determine if this protein was a substrate of the T7SS and if so to figure out its biological function.
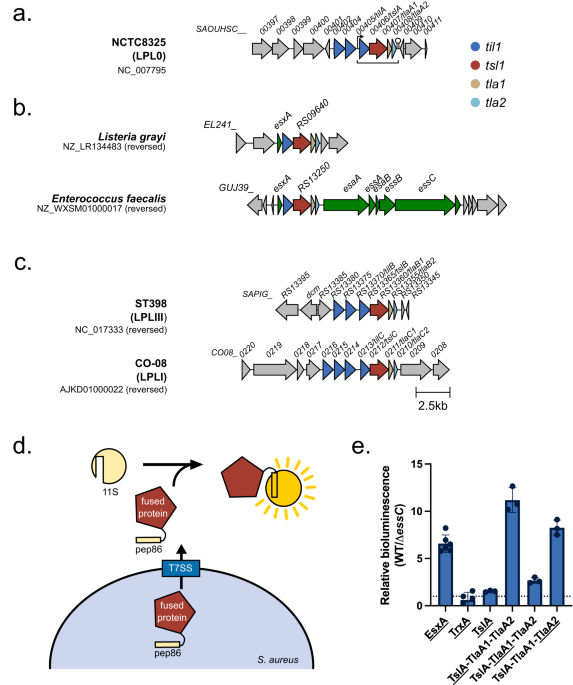
I joined the Palmer lab in late 2019 and so had little time at the bench before the global pandemic struck. This time spent working from home gave me the opportunity to do some bioinformatics analysis on this putative T7SS substrate. Prediction software identified potential similarity in the N-terminal region to known lipases, leading us to call this putative substrate, Type VII Secreted Lipase A (TslA). However, all T7SS toxin substrates described to date carry a helical LXG domain at the N-terminus, required for targeting the protein to the secretion system. Whilst this was odd, further analysis identified a helical domain at the C-terminus of TslA. Furthermore, I noted that two small helix-turn-helix proteins were encoded downstream of tslA at the same locus. As my PhD progressed, evidence emerged that small helical partner proteins were responsible for binding to the LXG domains of canonical T7SS toxins, to facilitate their secretion2,3. This raised the possibility that the small helical proteins encoded adjacent to TslA may play a similar role.
Unfortunately, when I tested the T7SS-dependent secretion of TslA I was unable to reliably detect the protein in culture supernatants by western blotting, most likely due to poor secretion activity in lab growth media. Fortuitously, at the same time another PhD student in the lab, Yaping Yang, was developing a highly sensitive secretion assay using NanoLuc luciferase technology4,5. Using Yaping’s methodology allowed me to demonstrate that TslA is secreted in a T7SS-dependent manner, and that its secretion was dependent on the presence of the two small helical proteins, which we subsequently renamed TlaA1 and TlaA2 (Type VII Secreted Lipase Associated Protein A1 and A2).
We formulated a hypothesis that TslA was used to kill other bacteria due to the presence of a cluster of non-identical lipoproteins encoded upstream of tslA, which we thought might be immunity proteins required to block TslA lipase activity. These lipoprotein genes, present in variable copy number across S. aureus strains, were shown to recombine at high frequency to produce new variants, as has been observed for other T7SS-associated immunity genes6–8. We therefore named these lipoproteins as the Type VII Immunity against Lipase family 1 (Til1). My bioinformatic analysis had identified that these lipoprotein gene clusters could be found at four loci in the S. aureus chromosome and that TslA homologues could be encoded at up to three of these, in a strain-dependent manner. To determine if TslA was toxic to the cell, it was necessary to delete these til1 loci. However, with 18 of these genes encoded in the genome of our lab strain USA300, this was going to be a tall order. Fortunately, Prof. Simon Heilbronner (University of Tübingen) had already made such a strain8, which he very kindly shared with us. Using this strain allowed me to confirm that TslA was toxic to the producing cell in the absence of these Til1 proteins, but only when it could be secreted from cell by the T7SS. Moreover, I was also able to show that the Til1 protein encoded directly adjacent to tslA was sufficient to fully rescue this toxicity when the encoding gene was reintroduced onto the chromosome, confirming that this is the immunity protein.
Once I knew that TslA was a T7SS substrate that could kill the producing cell from the outside if Til1 immunity proteins were lacking, I wanted to determine the source of this toxicity. Dr Nicole Mietrach in the Palmer group was able to demonstrate that TslA had lipase activity against a model substrate, and that this activity was inhibited by binding of the immunity protein, TilA. To determine the specify of this lipase, Prof. Terry Smith (University of St Andrews) kindly performed lipidomics to identify the target of TslA. TslA was found to have activity against a range of lipids in the S. aureus membrane, degrading these through indiscriminate phospholipase A activity. By removing a single acyl chain from phospholipids, this has a detergent-like effect in the membrane, results in significant membrane damage and ultimately cell death.
Satisfyingly, I have been able to fulfil the aims of my PhD, showing that indeed TslA is a substrate of the T7SS and that it has antibacterial lipase activity. TslA is the first example of a T7SS substrate with a targeting domain present at the C-terminus rather than the N-terminus. This is quite a surprising finding, as it is unusual for a protein translocation machinery to be able to recognise a secretion signal at either end of a substrate protein. In future it would be interesting to understand the features of both the substrates and the secretion system that facilitate this unusual dual recognition.
References
- Ulhuq, F. R. et al. A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc. Natl. Acad. Sci. U.S.A. 117, 20836–20847 (2020).
- Klein, T. A. et al. Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System. mBio 13, e02137-22 (2022).
- Yang, Y. et al. Three small partner proteins facilitate the type VII-dependent secretion of an antibacterial nuclease. mBio e02100-23 (2023) doi:10.1128/mbio.02100-23.
- Yang, Y., Alcock, F., Kneuper, H. & Palmer, T. A high throughput assay to measure Type VII secretion in Staphylococcus aureus. http://biorxiv.org/lookup/doi/10.1101/2023.06.03.543475 (2023) doi:10.1101/2023.06.03.543475.
- Dixon, A. S. et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 11, 400–408 (2016).
- Tsuru, T. & Kobayashi, I. Multiple Genome Comparison within a Bacterial Species Reveals a Unit of Evolution Spanning Two Adjacent Genes in a Tandem Paralog Cluster. Molecular Biology and Evolution 25, 2457–2473 (2008).
- Garrett, S. R., Mariano, G., Dicks, J. & Palmer, T. Homologous recombination between tandem paralogues drives evolution of a subset of type VII secretion system immunity genes in firmicute bacteria. Microbial Genomics 8, (2022).
- Belikova, D., Jochim, A., Power, J., Holden, M. T. G. & Heilbronner, S. “Gene accordions” cause genotypic and phenotypic heterogeneity in clonal populations of Staphylococcus aureus. Nat Commun 11, 3526 (2020).
Read the article here: https://doi.org/10.1038/s41467-023-44221-y
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in