Early-life microbiota diversification facilitates enterocyte proliferation
Published in Microbiology

The adult gastrointestinal tract is colonized by trillions of bacteria that influence cellular homeostasis through microbial metabolism of dietary and hepatic compounds. The microbiota ecosystem and associated metabolic networks are gradually established in the first years of life (2-3 years) beginning at birth. In healthy term infants, lactic-acid producing bacteria typically colonize the gut shortly after or during delivery and gradually increase in abundance with limited diversity throughout infancy. Delivery at earlier gestational ages (pre-term) is associated with expedited microbiota diversification and altered metabolic profiles in the gut. Evidence from germ-free mouse models has shown that microbial acquisition in the gut facilitates tissue growth and organization of the crypt-villus architecture associated with a mature intestinal tract. However, it remains unclear if distinct patterns of term and pre-term microbiome maturation confer functional changes in the developing small intestine.
Two neonatal intensive care unit physicians, Dr. Oleksandr Kudin and Dr. Josef Neu had access to fecal samples from term and pre-term patients and shared our interest in elucidating host response to neonatal microbiota. Dr. Kudin joined our research group as part of his research fellowship, and together we optimized organoid cultures for in vitro screening of microbial metabolites from patient fecal samples. Simultaneously, other publications demonstrated that lactate (derived from lactic acid producing bacteria in the term gut or from ingested milk) induces enterocyte proliferation, while structural proteins from bacteria (lipopolysaccharide, flagellin) activate toll-like receptor signaling and inflammatory responses. Given this evidence, we expected pre-term microbial communities to induce an inflammatory response while term microbial metabolites would facilitate epithelial maturation.
We first characterized microbial and metabolic composition in term and pre-term cohorts. As expected, term samples harbored microbial communities characterized primarily by lactic-acid producing bacteria (LAB). In contrast, the pre-term microbiome was more diverse and lacking prominent LAB species. The pre-term cohort exhibited a significantly higher metabolic capacity, notably including significant increases in secondary bile acids. These biotransformed bile acids have been shown to increase proliferation and epithelial regeneration following chemical-induced injury. Mapping metabolomics profiles to microbial KEGG pathways showed a significant increase in amino acid and vitamin biosynthetic pathways with a concomitant increase in overall metabolite concentrations.
To examine how these metabolites functionally affect the intestinal epithelium, we cultured small intestinal organoids with term and pre-term fecal metabolites. These organoids form “mini-guts” derived from intestinal stem cells which recapitulate epithelial morphology and cellular heterogeneity. Surprisingly, when cultured in the presence of pre-term fecal metabolites, we observed increased proliferation (fig. 1) and stem cell activation relative to term fecal metabolites without activation of inflammatory signaling pathways.
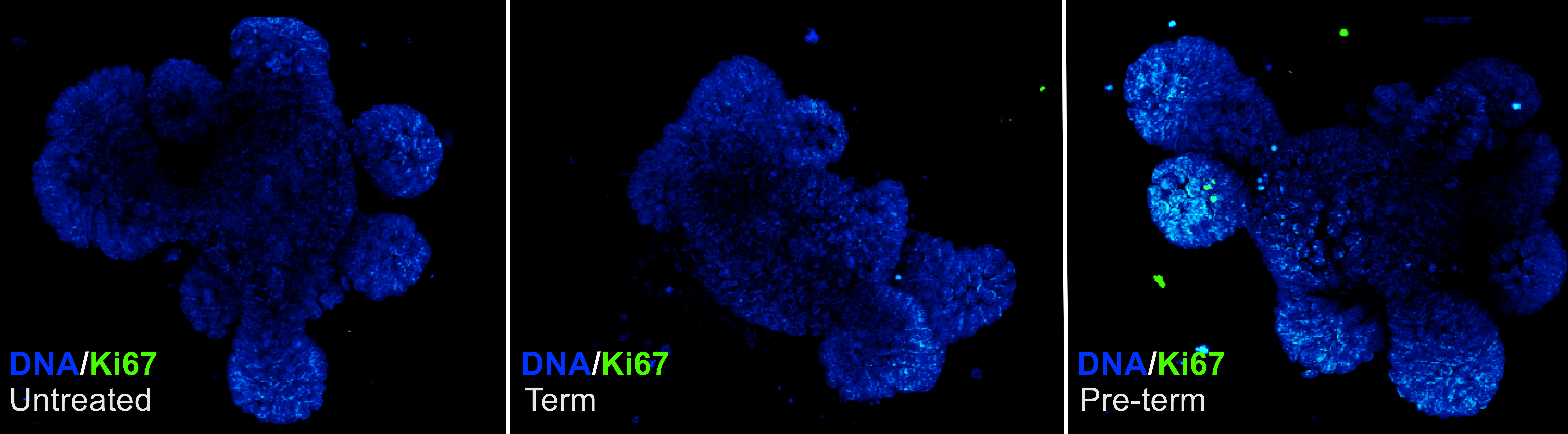
Organoid models are an increasingly common and powerful tool for defining host-microbe interactions intrinsically linked to the biology of the intestinal epithelium. Recently, human organoids have been used to identify the functional effects of individual microbial metabolites and characterize mutational profiles after exposure to bacterial genotoxins. Our study demonstrates the capability to induce functional changes associated with entire microbial ecosystems in vitro using host fecal material. This approach may help clarify complex metabolic interactions between host and microbes in intestinal diseases such as colon cancer.
For more details, please refer to our paper.
Follow the Topic
-
BMC Microbiology

This is an open access, peer-reviewed journal that considers articles on all microorganisms - bacteria, archaea, algae and fungi, viruses, unicellular parasites and helminths.
Related Collections
With Collections, you can get published faster and increase your visibility.
Current trends and future directions in mycology
Fungi are a diverse ubiquitous group of eukaryotic organisms, comprising e.g. unicellular yeasts as well as multicellular filamentous microorganisms and mushrooms. They exhibit remarkable morphological and ecological diversity and fulfil a wide array of biological and ecological roles as pathogens, decomposers, and mutualists. The field of mycology, dedicated to the study of fungi, has gained increasing importance in recent years, owing to both the beneficial and harmful impact of fungi on human health, agriculture, and the environment.
Recent advances in next generation sequencing, multi-Omics technologies, molecular biology and bioinformatics have significantly enhanced our understanding of the biology and ecology of fungi, as well as the complex interactions within fungal communities and their habitat. Research and innovation in fungal biotechnology have led to the development of antifungal agents, biocontrol methods, and the application of fungi in bioremediation and sustainable biofuel production. Meanwhile, research on fungal ecology has deepened our knowledge of the impact of fungi on ecosystem functioning and the implications of climate change on fungal diversity and distribution.
In support of United Nations’ Sustainable Development Goal 3 (SDG 3, Good Health and Well-Being), BMC Microbiology launches the Collection Current trends and future directions in mycology. This Collection invites contributions to current research and future perspectives in mycology, covering a wide range of topics e.g., related to the fungal ecological roles and response to environmental changes, as well as novel biotechnological applications of fungi. Research without a clear focus on fungi, fungal communities, or host-fungi interactions will not be considered. We invite researchers and experts in the field to submit research articles covering a broad range of topics, including, but not limited to:
Molecular mechanisms in the development and pathogenesis of fungi
Parasitic fungi as models to study host-pathogen interactions
Taxonomy and phylogeny of fungi, with particular interest in poorly studied habitats and extreme environments
Resolving species complexes and generic concepts of speciose fungal genera
The role of saprotrophic fungi in nutrient cycling and ecosystem functioning
Diversity and ecological significance of endophytic fungi for plant health
Ectomycorrhizal and arbuscular mycorrhizal fungi
Impact of climate change on the distribution and function of fungal communities
The potential of fungal secondary metabolites for novel drug discovery
Mechanisms of antifungal drug resistance in pathogenic fungi
Mycotoxins in food security
Applications of fungal enzymes for sustainable industrial applications
Mechanisms of mechanosensing and fungal contact sensing
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 24, 2026
The skin microbiome
The skin is the largest organ of the human body, colonized by a diverse and complex community of microorganisms, including bacteria, fungi, and viruses. The so-called ‘skin microbiome’ can affect human health, and its imbalance and dysbiosis can contribute to skin diseases. As a first primary protective barrier, the skin indeed plays a crucial role in human health by preventing the invasion of pathogens. Skin microbiome composition and associated diversity, depend on the microecology of the skin surface, available nutrient profile, and its topographical location in the body, as well as on various host-related and environmental factors. Moreover, the skin microbiota can be modulated by the cutaneous innate and adaptive immune responses, often resulting in dynamic interactions between skin-populating microorganisms.
Over the past decade, researchers have described the skin microbiome in unprecedented detail, revealing e.g. extensive communication between bacteria, skin cells and immune cells, and complex multispecies communities that affect skin health and immune responses. In support of United Nations’ Sustainable Development Goal 3 (SDG 3, Good Health and Well-Being), BMC Microbiology launches the collection ’The skin microbiome’. This collection aims to explore research on skin microbiome and their interactions with the human skin, the role of the skin microbiome in human health and disease, as well as novel approaches for the modulation, bioengineering or rebuilding of the skin microbiota. Research without a clear focus on skin microorganisms, host-microorganism interactions and/or microbiomes will not be considered. We invite researchers and experts in the field to submit research articles covering a broad range of topics including, but not limited to:
Interactions between microbiota and human skin
The role of skin microbiota in human health and disease
Skin microbiota colonization, assembly, diversity and ecology
Skin microbiome modulation by the skin immune system
The host skin barrier and microbial infection
Skin ageing and the skin microbiome
The prevention, diagnosis and treatment strategies for microbe-related skin conditions in health and disease
Multi-Omics approaches applied to skin microbiome research
Modulation of skin microbiota and skin-microbiota interactions using synthetic biology approaches
Modulation, manipulation and restoration of the skin microbiome in health and disease
The gut-brain-skin axis
Modeling the skin microbiome
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 07, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in