eDNA as a jawsome tool for ‘Seq’-ing sharks
Published in Ecology & Evolution

Who would have imagined that scientific concepts showcased in Crime Scene Investigation can potentially detect species that were once thought to be locally extinct? However, did you know that these molecular technologies can really be used to rediscover animals that have seemingly vanished, but are in fact leading private lives amongst us? As unimaginable as it sounds, the transformation of television fantasy to reality is now possible with recent environmental DNA (eDNA) tools. Akin to a crime scene scattered with clues (e.g., hair, fingerprints, and bodily fluids), the natural environment is chocked-full of trace DNA, be it from faeces or skin that have been released or shed by animals and plants that live in it. This trace genetic material, or eDNA, is detectable with molecular tools from a wide variety of environmental samples, ranging from seawater, soil, snow and even air. In the last five years, eDNA methods have become exponentially popular due to its broad applicability to address a wide range of scientific questions—from being hot on the trail of invasive species, to beating the bushes for biomonitoring endangered and elusive species.
Just six years ago when I was an undergraduate, the technology was in its infancy with just a handful of ‘proof-of-concept’ studies from freshwater and temperate marine water samples. Having just taken the first few baby steps into research, this technology was completely new to me and it was then that I entertained the thought, “I wonder if eDNA works in Singapore’s highly sedimented and tropical marine waters? Perhaps we may find more than the eye can see.” I was amazed by the revolutionary possibility of just collecting and sequencing seawater to reveal the organismal composition without having to physically observe them. Fast forward three years later, I managed to develop the pioneering eDNA protocol for detecting marine animals in Singapore at the Reef Ecology Laboratory, National University of Singapore. One fine morning during breakfast, our research fellow, Benjamin Wainwright, approached Marc Chang and I to ask if we were interested in testing my protocol for evaluating the diversity of sharks here. As an avid recreational diver with a particular fascination with large pelagics, I found this proposal extremely intriguing, and saw this as a perfect opportunity to test our recently developed eDNA protocols on conservation-related applications. Funny how our lab conjures the most exciting project ideas over mealtimes!

We learnt from our collaborators, Zeehan Jaafar and Kelvin Lim, that there used to be a much higher diversity of sharks and rays, based on records kept at the Lee Kong Chian Natural History Museum, Singapore, as compared to recent citizen science sightings. In the past, there were large elasmobranchs like whale and bull sharks despite the tremendous amount of shipping traffic in our port waters. Encounters nowadays, are sadly rare and are mostly by chance. It really made me wonder if these big charismatic fishes were driven to local extinction because of our island-city’s rapid coastal urbanization, or if they are merely seeking refuge elsewhere? These constitute the dark diversity—species that have been historically reported and still exist in the greater surrounds of their known geographic ranges but are presently missing from a specific area. Or it could also be phantom diversity—extant species that are locally colonised but have become too rare to be detected by regular survey methods.
We decided to apply our established eDNA protocol to reassess the shark and ray diversity in Singapore, as detailed in our paper published in BMC Ecology and Evolution. The fundamental questions we asked were: Can eDNA reveal more elasmobranch diversity than visually observed? Will the sequencing data be able to help make better informed management decisions in the future? One of the critical components for eDNA’s success is the availability of a robust sequence database for matching of our eDNA sequences. These will allow meaningful inferences of species identities, and we chose two gene regions for this purpose – 12SV5 and COI, as these reference sequences were well-represented in global databases. Since elasmobranchs are expectedly rare, we collected almost 100 water samples from across 9 sites, so as to broaden our scope of detection.
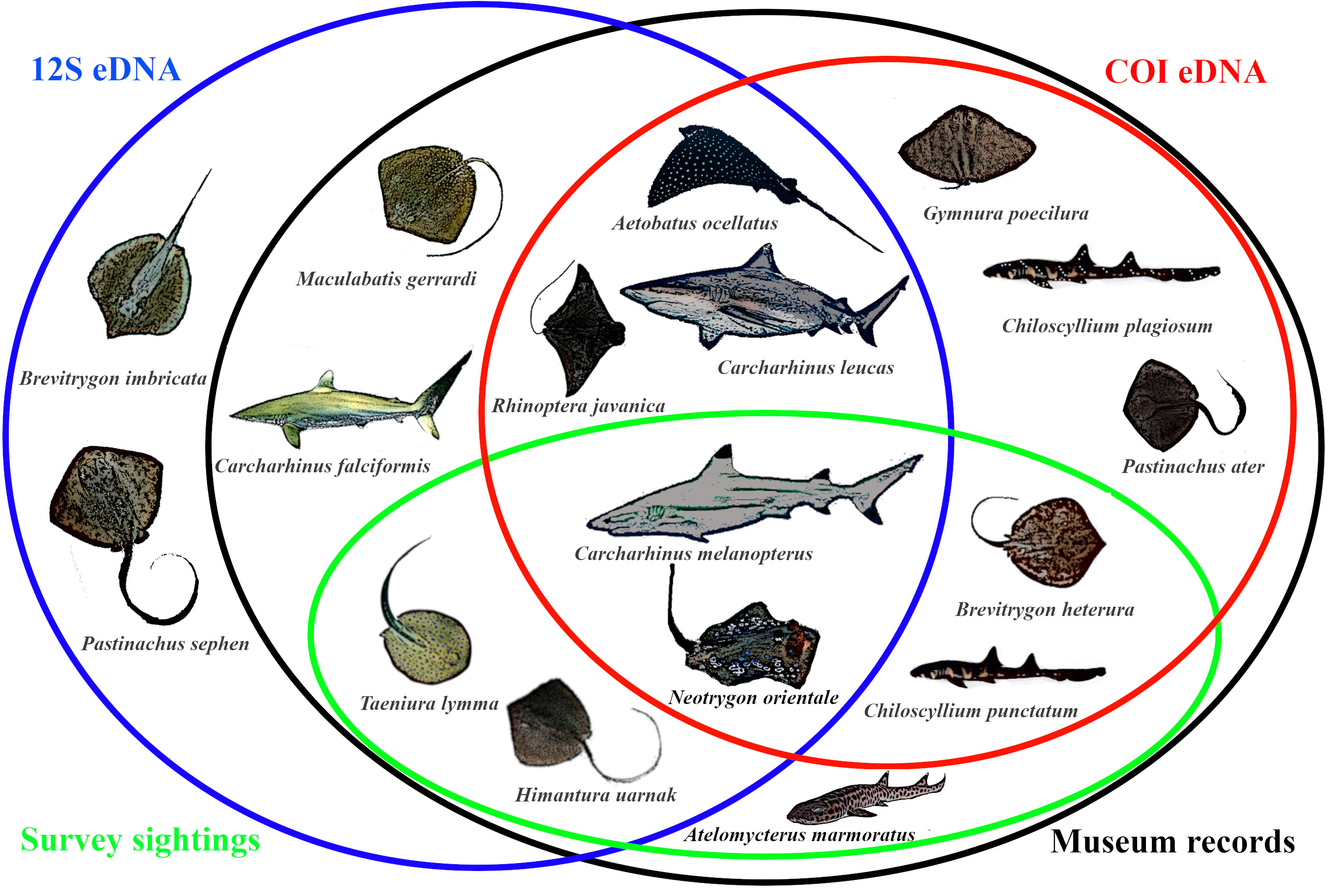
The most compelling result from our eDNA study was that we found up to 47 putative species, and managed to detect 16 sharks and rays with species-level identities. This is phenomenal, as eDNA found two times more species than the 7 (3 sharks and 4 rays) typically observed over the last twenty years. Comparing this updated species list from eDNA against the historical records, we found that the phantom diversity of sharks and rays have been considerably reduced by more than 20% after using eDNA. What is even more interesting is that our eDNA results uncovered two potential new species records! Of course, these records require further verification, possibly with evidence from sightings and proper identification of specimens by taxonomic experts. Nevertheless, the sensitivity of eDNA tools surpassing that of conventional sampling methods to uncover so much more elasmobranch diversity was extremely exciting for our team.
Our study has thus led to a deeper understanding of elasmobranch diversity in Singapore. We learned that the blacktip reef shark is extremely abundant, and there are still large body-sized species like bull sharks roaming our waters, albeit secretly. Further, we were able to make coarse relative abundance inferences of these species with normalised sequence read counts. We believe these will have implications for future work in conservation and formulation of management strategies.
It is increasingly apparent that eDNA metabarcoding methods hold enormous promise in revolutionizing the way we conduct species biomonitoring, community assessment and biodiversity conservation. With the rapid emergence of newer sequencing technologies, eDNA is now progressively applied to address a much wider array of different questions than ever before. Entering this exciting era of genomic research, we are highly encouraged by our study’s success in improved elasmobranch detection, and I feel extremely lucky to be able to conduct research work on a topic that I am passionate about. In the face of rising extinction rates, I look forward to continue using eDNA to complement conventional methods for improved species monitoring and to protect our natural heritage.
Follow the Topic
-
BMC Ecology and Evolution

An open access, peer-reviewed journal interested in all aspects of ecological and evolutionary biology.
Related Collections
With Collections, you can get published faster and increase your visibility.
Bioacoustics and soundscape ecology
BMC Ecology and Evolution welcomes submissions to its new Collection on Bioacoustics and soundscape ecology. By studying how animals use sound and how noise impacts them, you can learn a lot about the well-being of an ecosystem and the animals living there. In support of the United Nations Sustainable Development Goals (SDGs) 13: Climate action, 14: Life below water and 15: Life on land, the Collection will consider research on:
The use of sound for communication
The evolution of acoustic signals
The use of bioacoustics for taxonomy and systematics
The use of sound for biodiversity monitoring
The impacts of noise on animal development, behavior, sound production and reception
The effect of anthropogenic noise on the physiology, behavior and ecology of animals
Innovative technologies and methods to collect and analyze acoustic data to study animals and the health of ecosystems
Reviews and commentary articles are welcome following consultation with the Editor
(Jennifer.harman@springernature.com).
Publishing Model: Open Access
Deadline: Mar 27, 2026
Impact of climate change on ecology and evolution
BMC Ecology and Evolution is calling for submissions to our Collection on Impact of climate change on ecology and evolution. This Collection seeks to explore how climate change alters ecological dynamics and evolutionary processes, including shifts in phenology, local adaptations, and responses to invasive species. By understanding these shifts, we can gain insights into the resilience of ecosystems and the adaptive capacity of species in a rapidly changing world.
The significance of this research is underscored by the ongoing challenges posed by climate change, which threatens biodiversity and disrupts ecosystems. Recent advances in ecological modeling and genetic analyses have provided new tools to assess the impacts of environmental change on species and communities. These insights are crucial for developing conservation strategies and management practices aimed at mitigating the effects of climate change and preserving biodiversity for future generations.
Continued research in this area promises to enhance our understanding of the interplay between climate change and ecological dynamics. As new data emerges, we may uncover novel adaptive strategies employed by species in response to environmental shifts, revealing patterns of gene flow, population dispersal, and phenotypic plasticity. This knowledge can inform conservation strategies that are increasingly vital in an era of unprecedented environmental change.
•Climate change and biodiversity loss
•Phenotypic plasticity in response to environmental change
•Effects of invasive species on ecosystems
•Local adaptation and genetic structure in changing environments
This Collection supports and amplifies research related to SDG 13: Climate Action and SDG 15: Life on Land.
Publishing Model: Open Access
Deadline: Mar 03, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in