Efficient TnpB genome editors identified by a large-scale screening
Published in Bioengineering & Biotechnology

CRISPR-Cas technology has revolutionized biomedical research. Evolutionary classification of CRISPR-Cas systems is continuously being updated, with new subtypes being annotated and exploited for gene editing. Recent years, several CRISPR-Cas subtypes with more compact size have been identified, such as SaCas9 (1053 aa)1, Nme2Cas9(1082 aa)2, CjCas9 (984 aa)3, while their sizes are still close to 1000 aa, and the fusion of regulatory domain, base editor, or prime editor often exceeds the size limitation of AAV vector. Latest, a much smaller subtype, Cas12f systems (410-550 aa)4-7, have been identified. However, their editing frequency remains below 10% on average, even after extensive optimization. Therefore, novel miniature gene editors with high efficiency are highly desirable.
On a different note, the evolutionary origin of CRISPR-Cas has also been explored. In 2015, Eugene V. Koonin proposed that IscB and TnpB, derived from prokaryotic transposon family IS200/IS605, are likely ancestors of Cas9 and Cas12, respectively8. Building upon this notion, Zhang and Siksnys groups elucidated the function of IscB and TnpB, both working as an RNA-guided DNA endonuclease, in a manner similar to that of CRISPR-Cas system9, 10. Both IscB and TnpB proteins are exceptional compact (350~550 amnio acids long). In comparison to IscB, TnpB are much more widespread with over one million putative loci in bacteria and archaea genomes. Therefore, TnpB represents an enormous resource of miniature genome editors. However, the functional and evolutionary characteristics of TnpB systems are largely unknown and large-scale in silico analysis and experimental screening strategy remains to be developed.
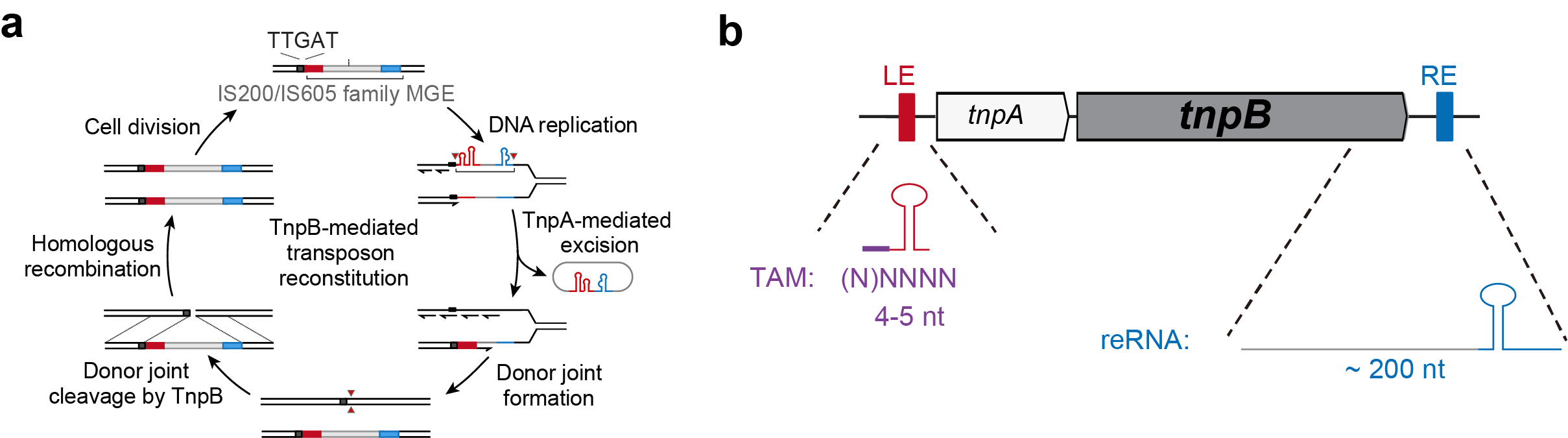
Fig. 1 | Analysis of TAM and reRNA sequence of TnpB proteins. a, Proposed role of TnpB in transposition. Panel a adapted from Karvelis et al [10]. b, A schematic of IS605 transposon. The genomic position of potential TAM and reRNA sequence of TnpB editors are labelled.
Inspired by the “Peel, Paste and Copy” model proposed by Siksnys et al.10, we hypothesized that both TAM sequence and reRNA backbone could be directly retrieved from the genomic context of each TnpB locus (Fig. 1a). We tested 64 candidates annotated in ISfinder, which is the most comprehensive database of insertion sequences. The results showed the preferred TAM sequence of each TnpB is generally identical to the cleavage sequence upstream of left end (CL, 4-5 nt) of each IS200/IS605 and the 3’ terminal of reRNA backbone exactly matches the right end (RE) of IS200/IS605 loci (Fig. 1b). Taken together, these results collectively indicate that TAM and reRNA backbone can be precisely retrieved from genomic sequence.
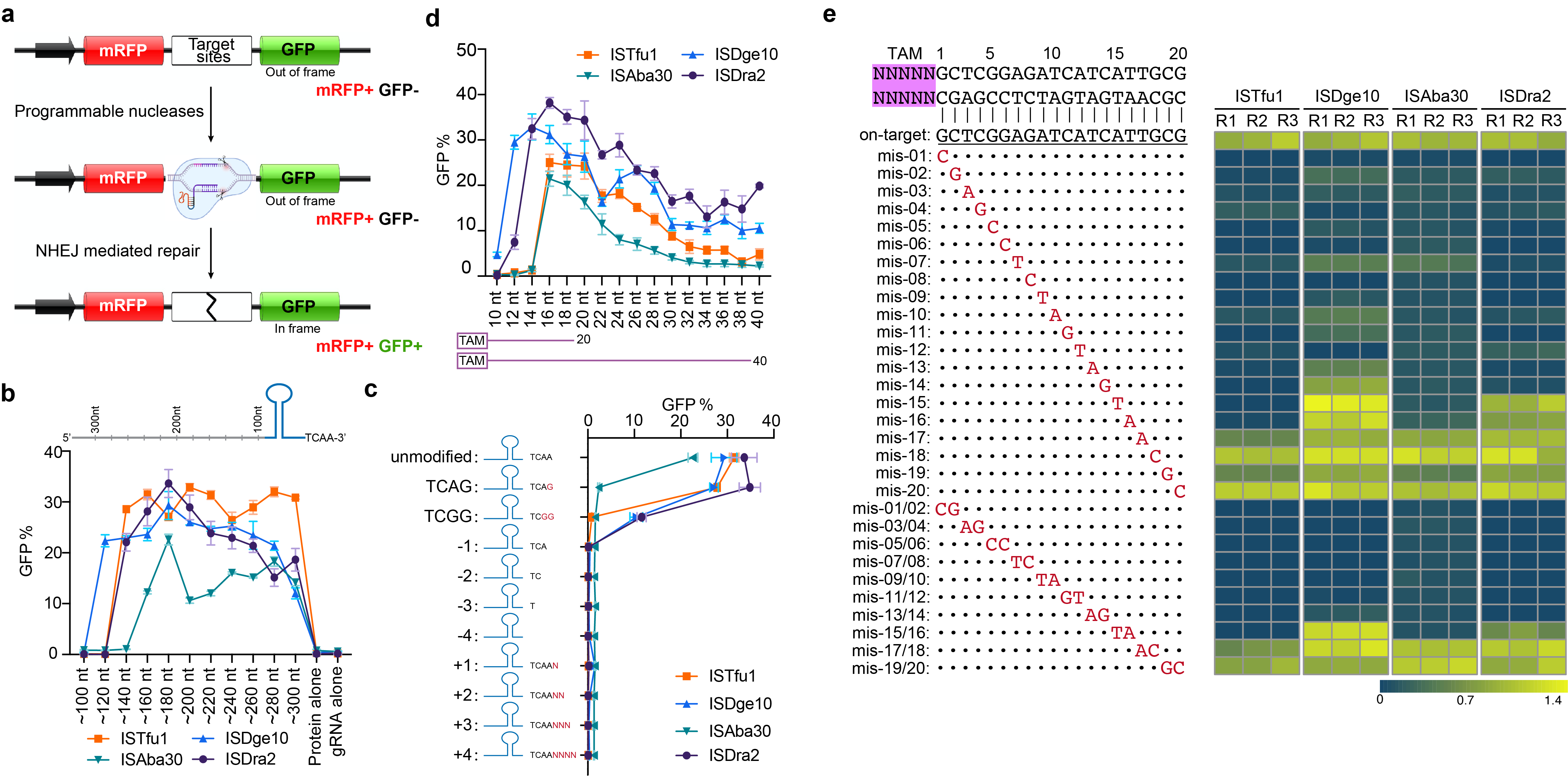
Fig. 2 | Characterization of TnpB-associated reRNA. a, A schematic showing the surrogate reporter. b, reRNA scaffold with 5’ sequences ended in different positions; c, reRNA scaffold harboring different 3’-end sequence contexts; d, guide segment of different lengths; e, guide segment with sequential one or two nt mismatches. Gene editing efficiency was quantified via the reporter assay, data are shown as the mean ± SD of three biological replicates.
We then characterized the crucial factors for reRNA function for TnpB proteins using surrogate fluorescent reporter and found: First, TnpB systems can tolerate a variety of reRNA scaffold sizes, with 120-300 nt scaffolds supporting editing activity. Second, 3’ terminal of reRNA must precisely match 3’ end of RE, adding or removing a single base at the 3’ end significantly reduces the activity. third, TnpB preferred 16-20 nt target length. Last, the minimal seed region of reRNA guides comprised the 12nt adjacent to TAM (Fig. 2). Altogether, the above characterization defines the key parameters for designing an efficient reRNA in the TnpB system.
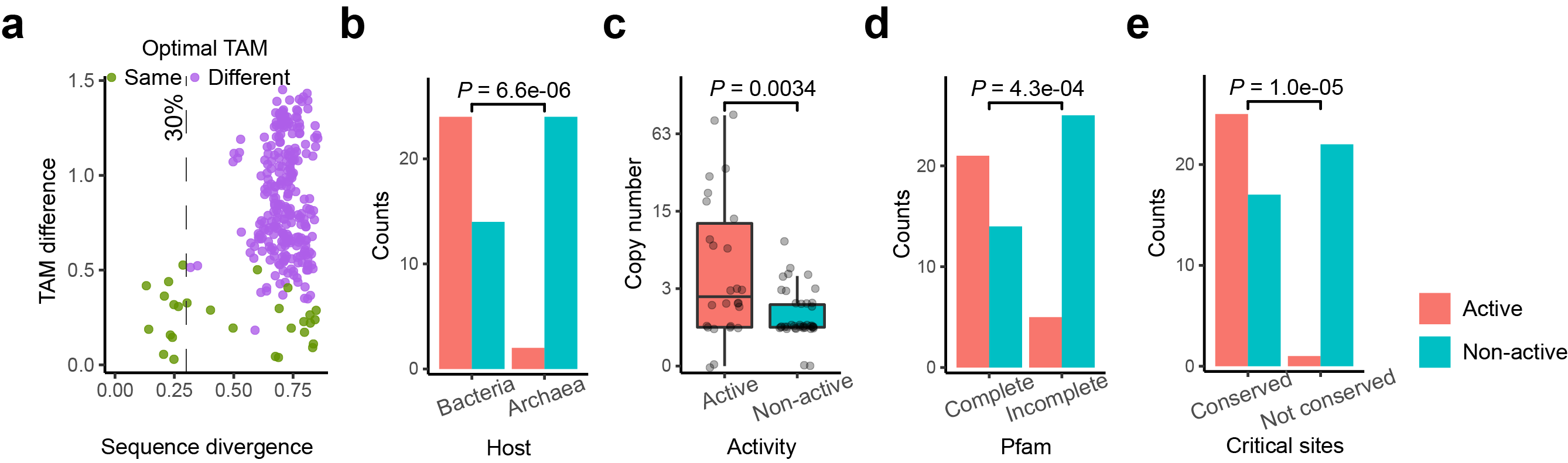
Fig. 3 | Functional properties of active TnpB systems. a, Pairwise comparison of active TnpB systems showed that similar (<30% divergence) TnpB proteins share the same optimal TAM sequences. The X-axis shows pairwise protein sequence divergences, while the Y-axis shows the distance of the corresponding pair of TAMs (motifs). b-e, The association between the presence/absence of TnpB-mediated genome editing activity in E. coli. and host, copy number, detection of 3 domains and presence of 10 conserved residues. For copy number, the box plot shows the median by black line, first and third quartiles by hinges, and minimum and maximum by whiskers. Wilcoxon test was performed (n = 26 vs. 39). For other factors, Fisher’s Exact tests were performed.
We next performed protein-centric analyses to differentiate active and inactive TnpB systems. Only 2 TnpB systems from archaea are active when transferred into E.coli, while the number increases to 24 for TnpB systems from bacteria. Since the copy number of tnpB loci reflects their transposition activity, active TnpB systems tend to have higher copy number. In addition, compared to inactive TnpBs, active TnpBs encode all three characteristic domains (HTH, OrfB_IS605, ZnF) at a higher proportion (81% vs. 36%). By examining the protein alignment, we identified 10 amino acids conserved across active TnpBs, which could be a criterion to select active ones (Fig. 3).
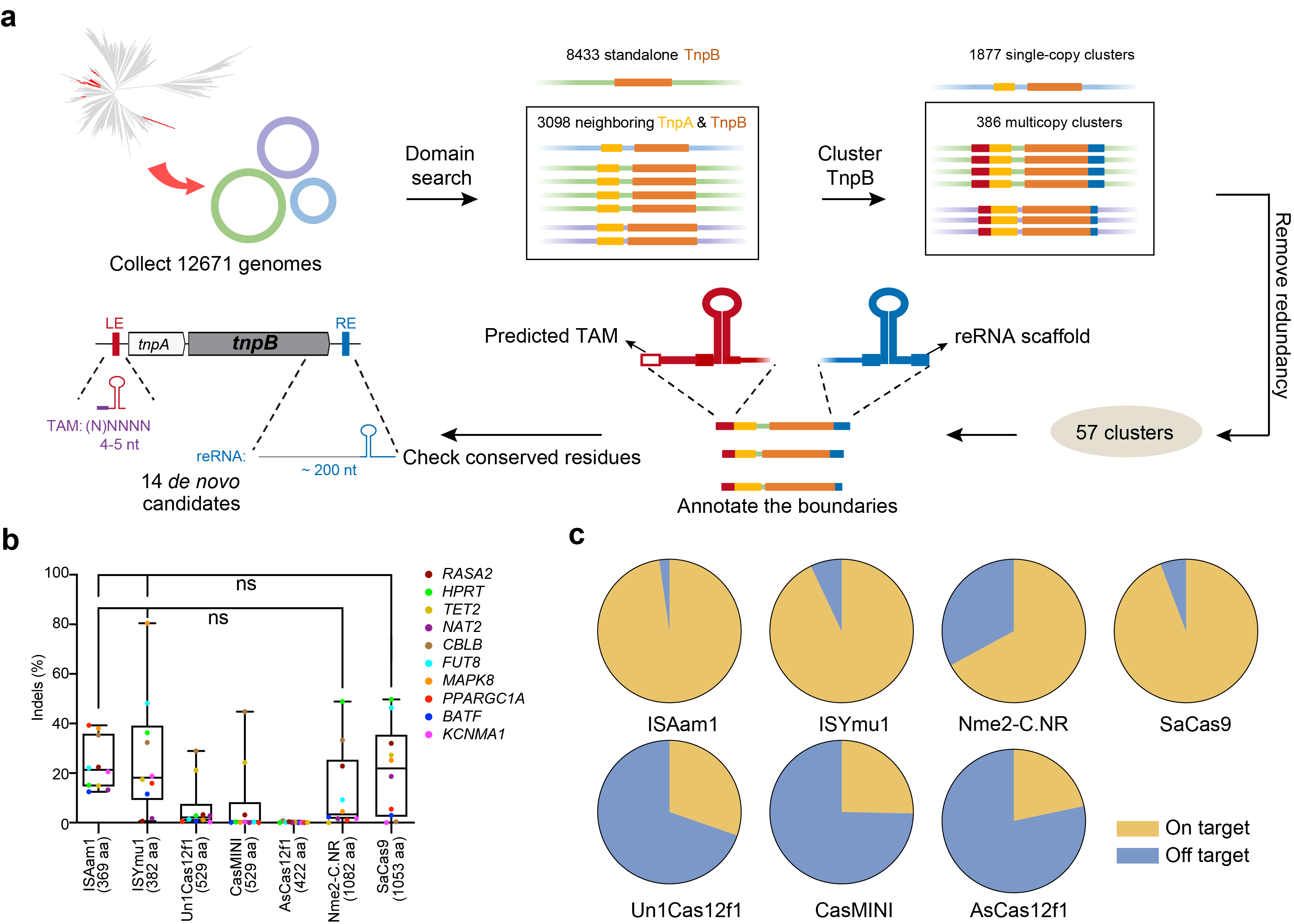
Fig. 4 | De novo annotation and characterization of ISAam1 and ISYmu1 TnpB systems. a, A schematic showing the pipeline for de novo annotation of TnpB systems. b, Comparison of editing efficiency of two TnpB systems and five Cas nucleases at 10 genomic loci in human HEK293T cells. Each dot represents the average efficiency of three biological replicates. Ordinary one-way ANOVA test with Tukey’s multiple comparisons was performed (**P < 0.01; *P < 0.05). c, The off-target level quantified by iGUIDE analysis at MAPK8 locus in HEK293T cells. The pie chart shows the proportion of on-target and off-target reads, respectively.
Followed the route we established, we developed a de novo annotation pipeline and chose 14 potential candidates for functional screen. Among these, two (ISAam1 and ISYmu1) showed high gene editing activity in mammalian cells. To perform an in-depth evaluation of the application potential of ISAam1 and ISYmu1 TnpBs, we examined their activity together with specificity in comparison with the five well developed small CRISPR-Cas editors, which include three Cas12fs, Nme2Cas9 and SaCas9. All gRNAs for each system were designed to overlap within a narrow range . The results showed ISAam1 and ISYmu1 TnpBs together with SaCas9 show relatively high activity and specificity than remaining systems (Fig. 4).
There are several exciting future directions to expand the research on TnpB systems, particularly considering their compactness, to further mine the IS1341 transposon family, to elucidate the mechanism of IS607 family and eukaryotic TnpB (Fanzor), to broaden targeting scope and to establish dead TnpB tools by fusing to functional elements, like DNA methylation domains. In Summary, TnpB systems as miniature gene editors present great potential for gene therapy.
References:
- Ran, F.A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186-191 (2015).
- Edraki, A. et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol Cell 73, 714-726 e714 (2019).
- Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8, 14500 (2017).
- Bigelyte, G. et al. Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat Commun 12, 6191 (2021).
- Kim, D.Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol 40, 94-102 (2022).
- Wu, Z. et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol 17, 1132-1138 (2021).
- Xu, X. et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell 81, 4333-4345 e4334 (2021).
- Kapitonov, V.V., Makarova, K.S. & Koonin, E.V. ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs. J Bacteriol 198, 797-807 (2015).
- Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57-65 (2021).
- Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692-696 (2021).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in