Endosomal Barriers Meet Chloroquine-like Carriers
Published in Bioengineering & Biotechnology and Microbiology
In the story behind the paper, a structure-guided design of endosomolytic lipids via integrating chloroquine’s structural motifs into ionizable lipids leads to the discovery of chloroquine-mimicking lipids for efficient mRNA delivery and genome editing.
Why Chloroquine?
Chloroquine is a versatile small-molecule drug that has been used to treat malaria and autoimmune diseases such as rheumatoid arthritis and lupus erythematosus. It has long been recognized for its ability to enhance nucleic acid delivery in vitro by destabilizing endosomal membranes, a major barrier in drug delivery. This property makes it valuable for enhancing the delivery of therapeutics that rely on escaping endosomes to reach their intracellular targets. However, its application as a delivery enhancer has been limited to in vitro co-incubation assays.
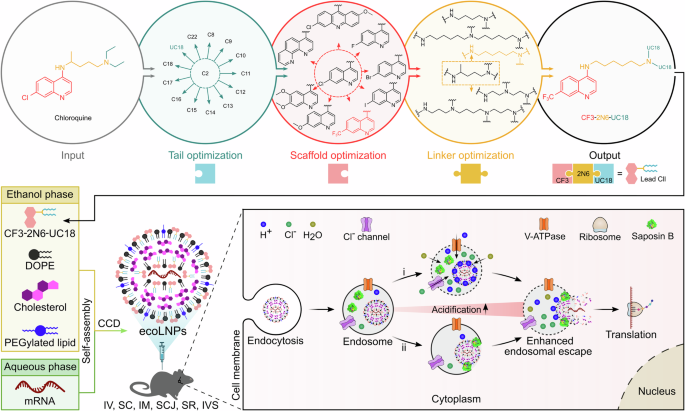
One of the major challenges with RNA delivery systems is ensuring the release of payloads from endosomes into the cytoplasm before degradation. Inspired by chloroquine’s endosomolytic property, we hypothesized that integrating its structural motifs into ionizable lipids could yield a new class of chloroquine-like lipid nanoparticles (LNPs) with enhanced endosomal escape capabilities. Our recently article published in Nature Communications demonstrates the feasibility of this integration strategy without using chloroquine itself. The newly developed mRNA delivery platform, termed endosomolytic chloroquine-like optimized lipid nanoparticles (ecoLNPs), facilitates endosomal escape primarily via the proton sponge effect and saposin B-promoted membrane disruption, ultimately enabling efficient mRNA delivery and genome editing.
Structure-Guided Modular Design of Chloroquine-like Lipids
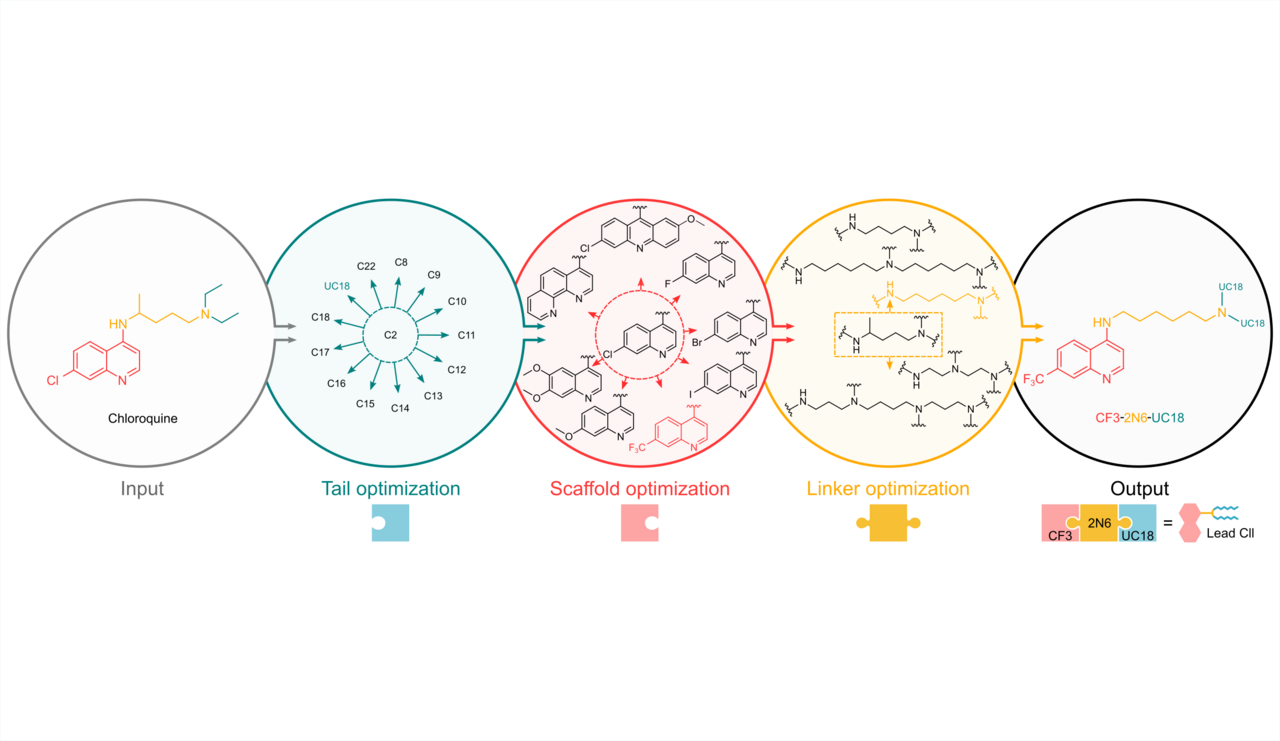
Since chloroquine itself is not a lipid, the first step was to create a lipid with chloroquine-like properties by incorporating signature structures extracted from chloroquine and ionizable lipids. We deconstructed chloroquine’s chemical structure into three modules and synthesized a combinatorial library of chloroquine-like lipids (Clls) via a triple combination of a quinoline scaffold, an ionizable linker, and a hydrophobic lipid tail. After multiple rounds of iterative screening and optimization using a high-throughput Central Composite Design method, we identified a lead Cll that exhibited up to 18.9-fold higher mRNA delivery efficiency than commercial transfection reagents.
Structure-Activity Relationship Analysis
Structure-activity relationship studies provide insights into designing highly active Clls: (1) Subtle variations in the length of carbon chain influence delivery potency of Clls. (2) Disruption of the quinoline scaffold by losing their signature quinoline rings impair Clls’ activity. (3) A trifluoromethyl substitution at the 7-position of the quinoline ring is highly favorable. (4) The introduction of excess ionizable nitrogen atoms linked with lipid tails may negatively impact delivery. (5) The quinoline scaffold, ionizable linker, and lipid tail of Clls synergistically affect their delivery performance.
Versatile Delivery Performance
On the one hand, ecoLNPs can mediate efficient delivery of plasmid DNA, siRNA, and various mRNAs to diverse cell types, including hard-to-transfect 3D cells. On the other hand, ecoLNPs are suitable for multiple administration routes, including intravenous, subcutaneous, intramuscular, subconjunctival, and subretinal injection, as well as intravesical instillation. Importantly, ecoLNPs not only induce robust mRNA-encoded protein expression at the injection sites following intramuscular injection with potency equivalent to SM-102 LNPs (a delivery carrier used in the Moderna’s COVID-19 vaccine), but also show higher tropism (90.2%) for lymph nodes than SM-102 LNPs (58.1%). Furthermore, ecoLNPs mediate successful genome editing by delivering both Cre mRNA and CRISPR-Cas9 mRNA in transgenic mice.
Stability, Mechanism, and Safety
ecoLNPs are capable of maintaining their in vitro and in vivo potency for at least 8 days after storage at 4°C, overcoming challenges related to ultra-cold storage and transportation. mRNA release from endosomes is confirmed by colocalization with endosomes and the calcein leakage assay. The robust endosomal escape and high delivery efficiency of ecoLNPs are attributed to two key mechanisms: the proton sponge effect and saposin B-mediated membrane disruption. Hemolysis assays confirmed their minimal toxicity at physiological pH but strong endosomal membrane disruption at endosomal pH. Safety evaluations in mice revealed no significant adverse effects on liver and kidney function or histopathology.
In summary, we develop chloroquine-like carriers mimicking the endosomolytic effect of chloroquine but without using the compound itself to achieve efficient mRNA delivery and genome editing. We will continue to focus on further iterations of this platform and explore their potential applications in mRNA vaccines, protein replacement therapies, cancer immunotherapy, and genome editing.
For further details, please see the full paper published in Nature Communications. Liu, Z., Wu, J., Wang, N., Lin, Y., Song, R., Zhang, M., Li, B. Structure-guided design of endosomolytic chloroquine-like lipid nanoparticles for mRNA delivery and genome editing. Nat Commun 16, 4241 (2025). https://doi.org/10.1038/s41467-025-59501-y
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in