Evolutionary plasticity of bacterial surface sugars
Published in Microbiology
Early in my scientific career, I became intrigued by the evolutionary problem of sexual reproduction, which more less goes as follows. If sexual reproduction entails gene shuffling, why is it so common if such shuffling can as much create adaptive gene combinations as it can destroy them? One of the proposed explanations for this is called the Red Queen Hypothesis. It posits that gene shuffling makes sense during antagonistic co-evolution, like between hosts and parasites, as rapid generation of novel diversity helps each side keep up in a never-ending arms race. While the Red Queen Hypothesis as a universal explanation for the ubiquity of sex has been heavily disputed, elevated rates of evolution and recombination are often linked to genetic regions which are engaged in antagonistic interactions (like MHC or ABO genes in vertebrates).
Horizontal gene transfer, or HGT, is often thought of as the sex counterpart in bacteria. However, it was not until genetic sequencing became common that we have started to acquire a good understanding of which regions in bacterial genomes are both linked with antagonistic interactions and evolve rapidly. One fascinating example is a genetic region that synthesises bacterial surface polysaccharides: polysaccharide capsules and lipopolysaccharides. A few things about such surface polysaccharide loci (or SP loci), stand out. First, SPs engage in multiple interactions with other microorganisms, immune system of vertebrates or plants and bacteriophages, many of which are antagonistic. Second, SP loci are hyper-diverse as SP synthesis requires sugar-specific enzymes. As these enzymes are highly diverse themselves, this produces endless combinatoric ways of making a sugar polymer. Third, genomic studies of SP loci in many different bacterial species have found that these loci exhibit elevated rates of evolution and homologous recombination. Finally, the genetic architecture of SP loci is consistent across the bacterial kingdom: variable, sugar-specific middle produce diversity and conserved flanks of the locus facilitate horizontal gene transfer of entire loci between bacterial lineages.
One major knowledge gap so far has been that studies investigating SP locus evolution have usually been considering it from the point of view of a single – typically medically relevant – bacterial species. Hence, we have little understanding of how SP loci evolve across species and genera. This is important as capsules and LPS are now considered as potential targets of novel medical interventions like glycoconjugate vaccines, immunotherapy and phage therapy. Therefore, a good understanding of the evolutionary dynamics driving SP loci is important from the point of view of disease management, for example by helping us predict which serotypes should be included in a treatment to minimise the risk of bacterial adaptation or ecological disruption.
To answer these questions, I paired up with Professor Kathryn E. Holt, Dr Kelly L. Wyres and Ryan Wick from Monash University and Dr Florent Lassalle from Imperial College London to analyse the diversity and evolutionary dynamics of SP loci across tens of different bacterial genera. We chose to work on the bacterial order of Enterobacteriales as (i) it features a lot of publicly available genomes, many of which are non-obligate pathogens from multiple environments, and (ii) many species are known to share a similar SP locus genetics. We decided to scan 27,334 genomes from 45 bacterial genera from the NCBI repository for known and previously unknown SP loci. We focused on two SP locus regions known from Escherichia coli studies (we referred to them as cps and kps) and widely reported in many other species. We used conserved genes as markers for sequence similarity search and extracted everything with a pre-defined genetic architecture that was potentially novel. Here is what we found.
First of all, we found a huge reservoir of genetic diversity of SP loci across the entire bacterial order, both in terms of the number of locus structures themselves (2654 unique cps structures in 22 genera and 332 kps structures in 11 genera; "structure" refers to the combination of gene families in the locus) and thousands of identified locus gene families. In fact, when we examined what is the best predictor of the SP genetic diversity, we found that it is sampling rather than genomic diversity. This suggests that as we sample and sequence new bacterial isolates, particularly from novel environmental niches, many new, previously unseen carbohydrate structures of capsules and lipopolysaccharides will be discovered.
Second, the population structure of SP loci was quite distinct from the whole-genome population structure. For example, we found that SP loci sharing a highly similar gene content were often shared by distant bacterial taxa, and that highly diverse, unrelated locus clusters often featured in single bacterial lineage. We also found weak evidence for a correlation between gene content similarity of SP loci and whole genome similarity. These findings imply that, at least at the genus and species level, SP loci evolve mostly independently from whole genomes.
Third, we found that many of SP locus structures have been recently exchanged by HGT between different species and genera (see figure below). (Note that 'recent' in this case has an evolutionary, not epidemiological, meaning.) Conversely, we also found examples of SP locus structures for which there is evidence of absence of recent HGT, suggesting that SP locus structures can also be selectively maintained during bacterial evolution. The likelihood of recently exchanging a SP locus via HGT varied significantly between different species, meaning that HGT likely has different impact on the SP locus diversity in different bacterial species. However, this implies that SPs during their evolution often cross species and genus barriers by HGT.
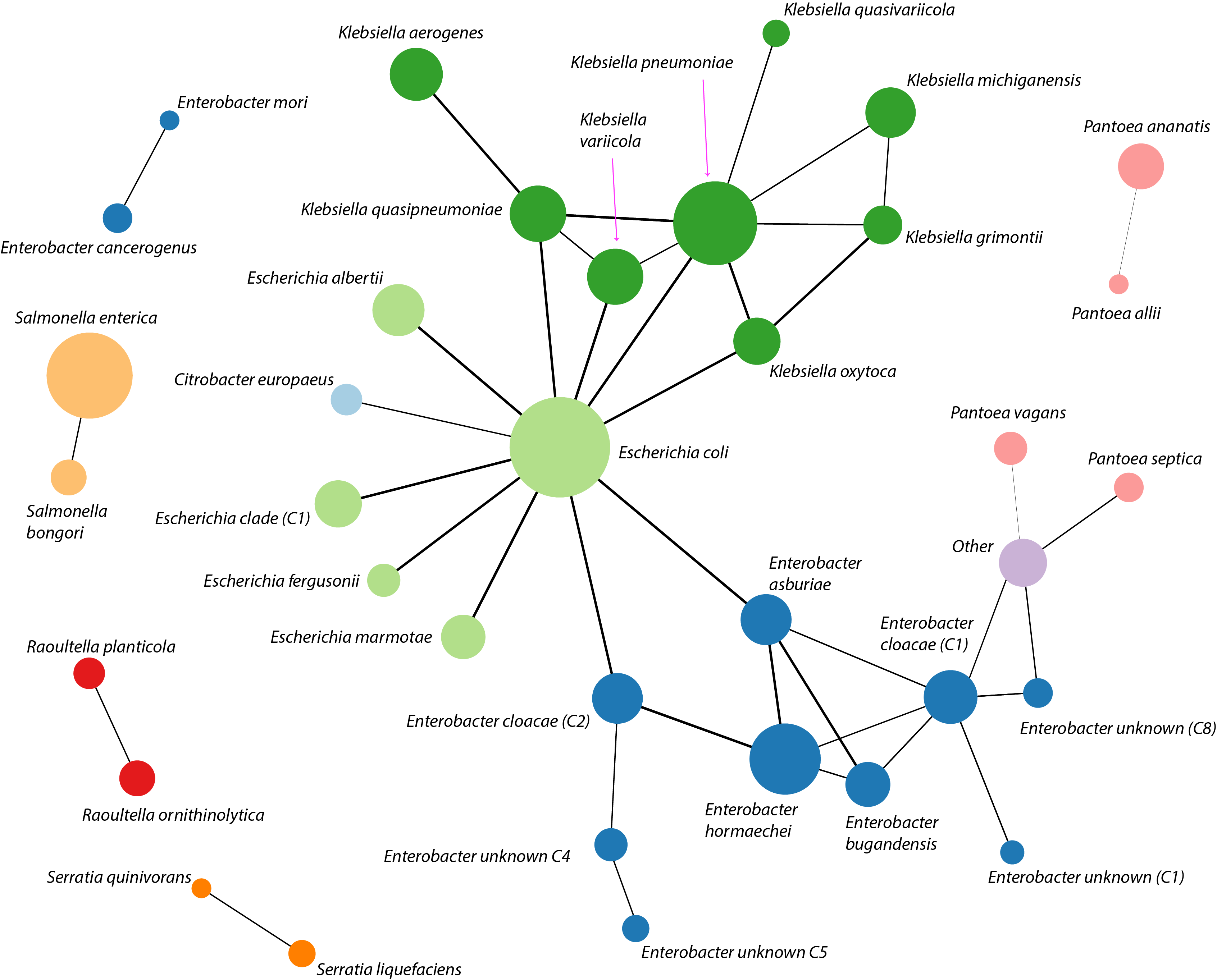
Figure. Impact of HGT on exchange of bacterial surface polysaccharides (SPs) between species and genera in. Each node of the network shows a species and edges link those species which were found to have exchanged a SP locus via recent HGT. Colour denotes a genus and node size reflects the sample size. Results only for the cps locus region shown.
Finally, in closely related groups of bacterial genomes (lineage-like clusters) we found relatively frequent movements of gene gain and gene loss within SP loci, often driven by individual gene movements. This would suggest that SP loci can evolve relatively quickly, sometimes even on epidemiological scales, emphasising the ‘plug-and-play’ nature of SP genetics.
While our study provides an unprecedented look at SP evolution in bacteria, it has two major areas of limitation. First of all, a genetic view on SP evolution is insufficient to properly inform us about different forms of selection shaping SPs in bacteria (e.g, converging selection) as well as about the SP genotype-phenotype map. Second, the epidemiological context of large genome database generation driving public repositories like NCBI makes it very difficult to reliably address various hypotheses about the interplay of ecology and evolution in shaping SP repertoire in bacteria.
Overall, our results demonstrate evolutionary plasticity of SP loci in bacteria and emphasise the range of different evolutionary forces shaping their repertoire. As of today we do not yet know the exact interplay between such plasticity and antagonistic interactions that bacteria engage in, for example via co-evolution with bacteriophages or host immune systems. However, I anticipate that bacterial capsules and lipopolysaccharides provide a wonderful context to study such interplays.
You can read the full story here: https://rdcu.be/b3p0a
You can also read about the research of this study's authors here:



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
López-Pérez M, Martin-Cuadrado AB, Rodriguez-Valera F. Homologous recombination is involved in the diversity of replacement flexible genomic islands in aquatic prokaryotes. Front Genet. 2014;5:147. Published 2014 May 22. doi:10.3389/fgene.2014.00147
Rodriguez-Valera F, Martin-Cuadrado AB, López-Pérez M. Flexible genomic islands as drivers of genome evolution. Curr Opin Microbiol. 2016;31:154–160. doi:10.1016/j.mib.2016.03.014