Expansion of Human Megakaryocyte-biased Hematopoietic Stem Cells by Biomimetic Microniche
Published in Healthcare & Nursing
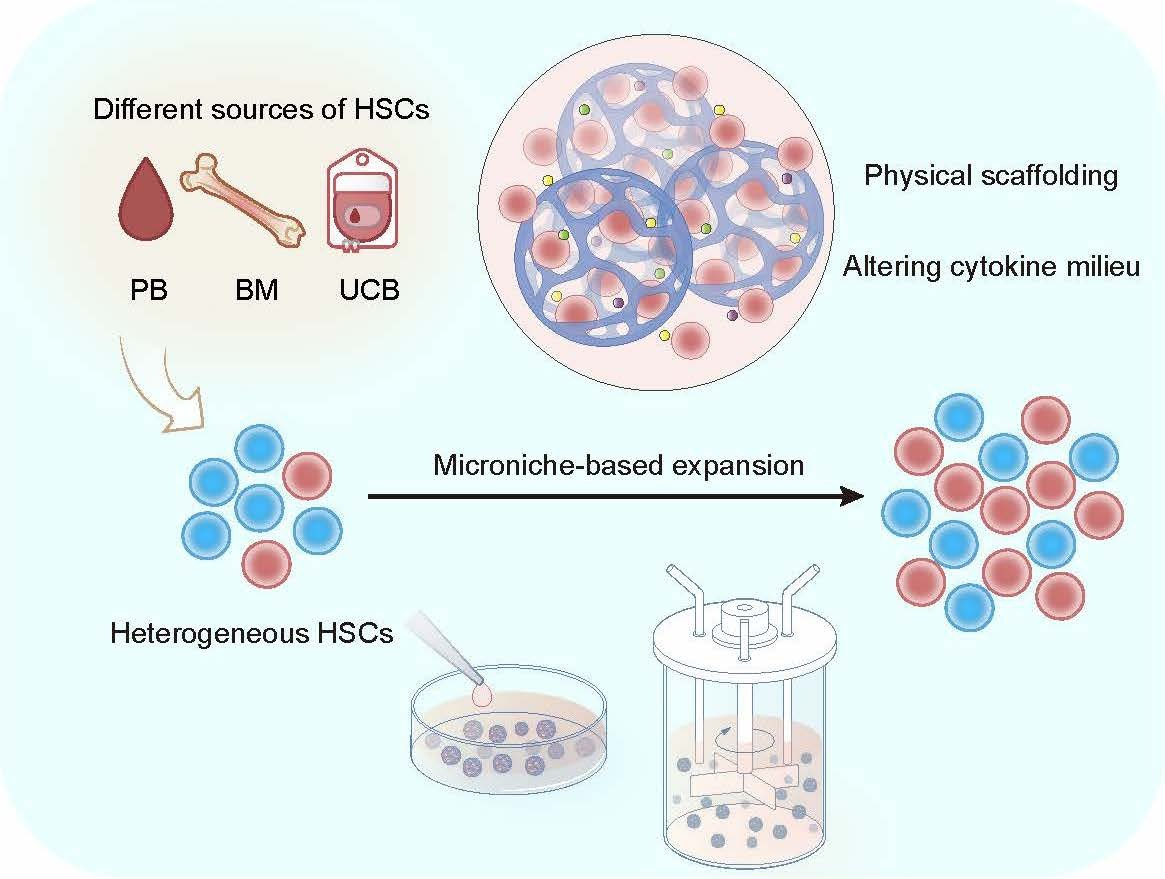
Hematopoietic stem cells (HSCs) are considered to be the basis of HSC-based cell therapy, but the main obstacles limiting their clinical application are the insufficient number of HSCs, and the failure to reconstitute specific lineages, especially megakaryocytes, after transplantation due to the functional defects of HSCs[1]. HSCs are a highly heterogeneous population in terms of biological function, molecular level, and cell fate. More and more evidence has confirmed the existence of lineage-biased differentiation and lineage-restricted HSC subsets, which have constantly revised and supplemented the classic balanced hematopoietic hierarchy[2]. Studies have demonstrated that the mice long-term platelet-biased HSCs reside at the top of the hematopoietic hierarchy[3, 4], while under pathological conditions there are subpopulations of human stem cells with a bias toward megakaryocytes[5, 6]. Under normal physiological conditions, human megakaryocytes can be directly derived from stem and progenitor cells in multipotential stage[7], suggesting that there may be megakaryocyte (Mk)-biased HSCs in human hematopoietic system, and loss or impaired function of this specific subpopulation may be responsible for the difficulties in reconstitution of the megakaryocytes. Unraveling these mechanisms and developing methods to expand Mk-biased HSCs may be a breakthrough to overcome associated reconstitution obstacles after chemoradiotherapy or hematopoietic stem cell transplantation (HSCT).
This study established a human HSC expansion strategy based on a biomimetic Microniche and realized the in vitro expansion of HSCs from different sources. It also revealed the existence of human Mk-biased HSCs and analyzed the immune phenotype of this subpopulation, proposing a new perspective for the heterogeneity and in vitro expansion of HSCs.
The study developed a 3D microniche that mimicked the bone marrow microenvironment in terms of rigidity, pore size, and materials, and added it to the cell culture system, and found that the system was able to expand primitive CD34+CD38-CD45RA-CD90+ and CD34+CD38-CD45RA-CD90+CD49f+ HSCs from a different sources including umbilical cord blood (UCB) CD34+ cells, as well as UCB, peripheral blood (PB) and bone marrow (BM) mononuclear cells (MNCs). According to the limiting dilution transplantation experiment, Microniche expanded the functional HSCs by 5.01-fold. BM MNCs from aplastic anemia (AA) patients cultured in this system could restore the lost hematopoietic reconstitution ability in immunodeficient mice. Next, after applying this expansion strategy in a bioreactor, the large-scale expansion of HSCs in vitro with UCB MNCs as the starting cell was realized, and 7.39-fold functional HSC expansion was reached.
Then, the study further investigated the effect of Microniche on megakaryotic hematopoiesis, and found that in the limiting dilution transplantation, the lymphoid (Ly) and myeloid (My) lineages of the successfully engrafted recipient were almost completely rebuilt, while only less than half of the recipient were able to reconstitute the Mk lineage, suggesting that there may be a previously unrecognized smaller subpopulation of cells within the heterogeneous pool of human HSCs with specific Mk reconstitution capacity, that is, Mk-biased HSCs. Fresh UCB HSCs that had not been cultured could reconstitute the Mk lineage in immunodeficient mice, but the mice in the control group could hardly reconstitute Mk. Limiting dilution analysis found that Microniche had expanded Mk-biased HSCs by 3.36-fold, which indicated that the Mk-biased HSC subpopulation was very sensitive in in vitro culture, and Microniche could maintain and expand the subpopulation that was original in physiological state and easily lost under traditional culture conditions.Single-cell sequencing found that the characteristic genes of the specific cell subpopulation after Microniche culture were SELL (CD62L) and PROM1 (CD133). Flow cytometry analysis also confirmed the existence of a unique cell subset of CD34+CD38-CD45RA-CD90+CD49flowCD62L-CD133+ in Microniche-based culture. By sorting this CD62L-CD133+ subpopulation and non-CD62L-CD133+ subpopulation for transplantation, it was confirmed that the CD62L-CD133+ subpopulation was responsible for the reconstitution of Mk, while non- CD62L-CD133+ subpopulation was responsible for other lineages (Figure 1). This shows that human Mk-biased HSCs are enriched in the CD62L-CD133+ subpopulation, and CD62L-CD133+ can be further used as cell surface markers to identify primitive Mk-biased HSCs immunophenotype based on long-term HSCs.
When designing strategies for expanding HSCs in vitro, it is necessary to fully consider the close two-way interaction between HSCs in the natural state and the microenvironment provided by physiological cell niches. Cells can perceive biochemical signals from the extracellular matrix (ECM), and continuously detect and respond to the mechanical, physical and mechanical characteristics of the external microenvironment; at the same time, cells actively change the characteristics of the microenvironment by secreting various factors and exerting traction to mechanically reshape the ECM, thereby generating complex cell-matrix mechanical interactions that play an important role in HSC homeostasis, regeneration, lineage choice, and migration[8-11]. This study found that the remodeling of the cytokine microenvironment and the rational physical support are critical for the expansion of human HSCs. The cytokine-cytokine receptors pathway was activated in the product cells of Microniche. Single-cell sequencing showed that there was a unique CD4-positive cell subpopulation in the product cells, which communicated extensively with other subpopulations through cytokine-cytokine receptors pathway. The comparison of various carriers with different rigidities, pore sizes and raw materials showed that Microniche appropriately simulated the physical structure of bone marrow cell niches, and more primitive HSCs and Mk-biased HSCs preferred to locate inside Microniche. This suggested that, based on the effective recapitulation of the biophysical and biochemical characteristics of the simulated bone marrow microenvironment, Microniche interacted with the HSCs, constructed a "small house" suitable for its own growth, and successfully maintained and expanded the Mk-biased HSCs. These results confirm the point made by previous study[12] that 3D culture that more closely simulates the environment of stem cell niches in vivo is beneficial to the "high-fidelity" expansion of HSCs in vitro, and this should be further investigated in related studies of HSC expansion. In the future, 3D microniche could be modified in vitro according to the unique ECM components and signaling molecules in the HSC microenvironment, so as to "tailor-made" biomimetic culture system suitable for different HSC subpopulations.
On the other hand, unlike previous studies that required the sorting of CD34+ or CD133+ cells as the starting cells for expansion, the microniche-based culture system can expand HSCs from MNCs obtained from human BM, PB or UCB (Figure 1), which means improved availability and convenience for clinical application in expanding HSCs for transplantation. Furthermore, microniche can be scaled up for the expansion of one or more UCB units in stirred bioreactors. The functions of the generated cells were also supported by xenograft experiments, which will also benefit the practical application of UCB HSCs in the future.
In conclusion, this study developed a biomimetic microniche-based human Mk-biased HSCs in vitro expansion technology, revealing the immunophenotype of human Mk-biased HSCs and the important role of biomimetic physical support for primitive HSCs. Meanwhile, a functional evaluation system for phenotype-defined HSC subsets was established, and the new megakaryocytes reconstitution in vivo evaluation strategy made up for the gaps in previous studies, improved the standard for stem cell function evaluation, and opened new possibilities for the establishment of clinical-grade HSC in vitro expansion strategies.
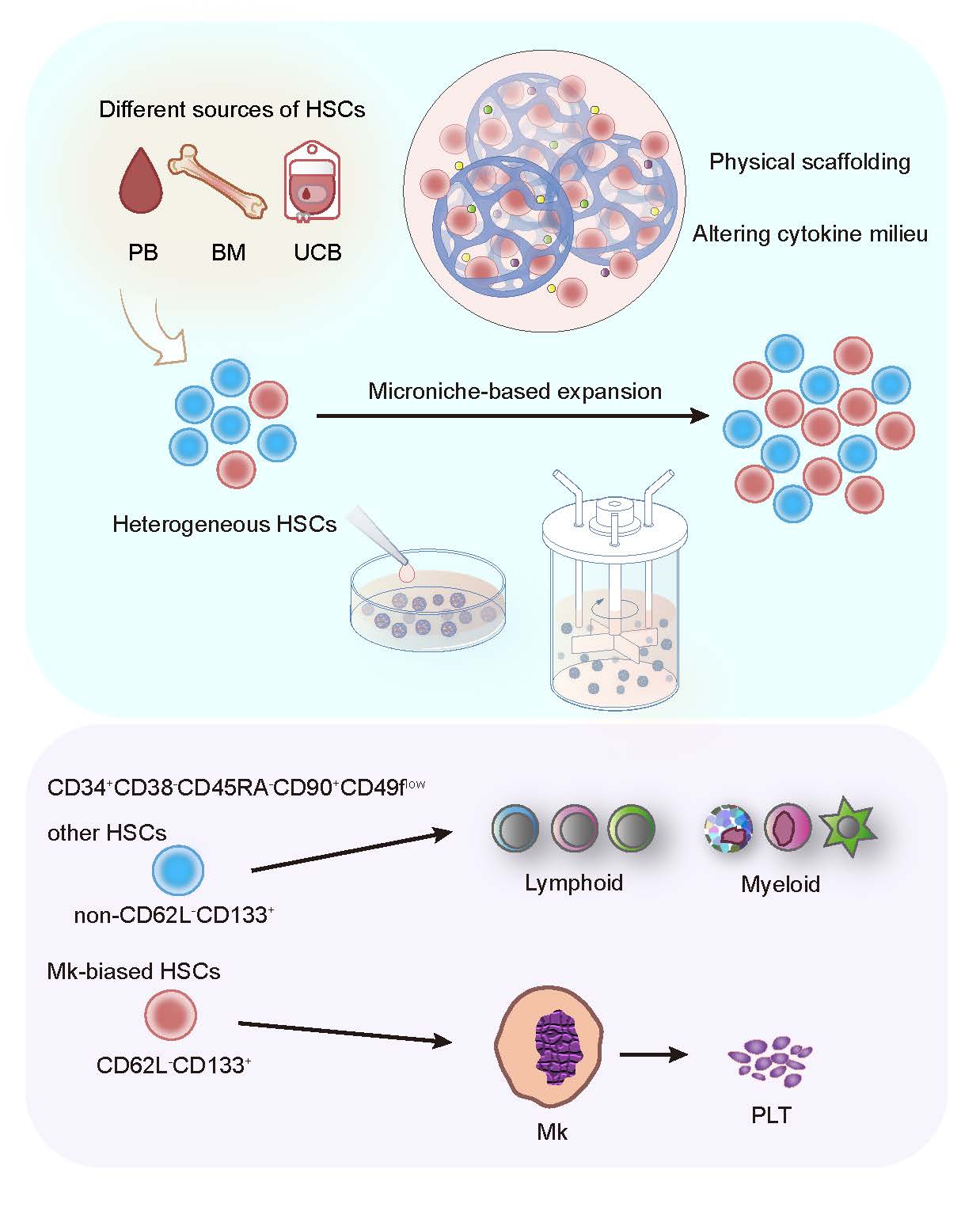
Figure 1. Expansion and identification of human megakaryocyte-biased hematopoietic stem cells by Microniche-based culture
References
[1] HUANG X, GUO B, CAPITANO M, et al. Past, present, and future efforts to enhance the efficacy of cord blood hematopoietic cell transplantation [J]. F1000Research, 2019, 8.
[2] ZHANG Y, GAO S, XIA J, et al. Hematopoietic Hierarchy - An Updated Roadmap [J]. Trends in cell biology, 2018, 28(12): 976-86.
[3] SANJUAN-PLA A, MACAULAY I C, JENSEN C T, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy [J]. Nature, 2013, 502(7470): 232-6.
[4] CARRELHA J, MENG Y, KETTYLE L M, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells [J]. Nature, 2018, 554(7690): 106-11.
[5] TONG J, SUN T, MA S, et al. Hematopoietic stem cell heterogeneity is linked to the initiation and therapeutic response of myeloproliferative neoplasms [J]. Cell Stem Cell, 2021, 28(4): 780.
[6] VAN EGEREN D, ESCABI J, NGUYEN M, et al. Reconstructing the Lineage Histories and Differentiation Trajectories of Individual Cancer Cells in Myeloproliferative Neoplasms [J]. Cell Stem Cell, 2021, 28(3): 514-23.e9.
[7] NOTTA F, ZANDI S, TAKAYAMA N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny [J]. Science, 2016, 351(6269): aab2116.
[8] VINING K H, MOONEY D J. Mechanical forces direct stem cell behaviour in development and regeneration [J]. Nat Rev Mol Cell Biol, 2017, 18(12): 728-42.
[9] LIU K, WIENDELS M, YUAN H, et al. Cell-matrix reciprocity in 3D culture models with nonlinear elasticity [J]. Bioact Mater, 2022, 9: 316-31.
[10] DAS R K, GOCHEVA V, HAMMINK R, et al. Stress-stiffening-mediated stem-cell commitment switch in soft responsive hydrogels [J]. Nat Mater, 2016, 15(3): 318-25.
[11] LIU K, VEENENDAAL T, WIENDELS M, et al. Synthetic Extracellular Matrices as a Toolbox to Tune Stem Cell Secretome [J]. ACS Appl Mater Interfaces, 2020, 12(51): 56723-30.
[12] Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel [J]. Nature Medicine, 2019.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in