Finding phages that infect bacteria with AI
Published in Microbiology, Computational Sciences, and Mathematics

Bacterial infections are an increasingly serious world problem
Bacteria are all around us. A lot of bacteria have positive effects on human health but others make us sick. For decades, we have successfully eradicated these disease-causing bacteria using antibiotics, saving millions of lives. Today however, bacteria are becoming increasingly resistant to most or all of the antibiotics we have available to fight them (Rossolini et al., 2014). A study from 2019 shows that resistant bacteria are directly responsible for an estimated 1.27 million deaths globally each year. What's worse, it is estimated that that number will only increase in the years to come if we do not take the appropriate measures and develop novel therapeutics.
Phages and phage therapy
Enter phages, the viruses that infect and kill bacteria. Phages were discovered in 1915 and 1917 by Frederick W Twort and Félix d'Herelle and by 1921 already they were first used therapeutically by researchers in Leuven, Belgium. Today, fueled by the declining effectiveness of antibiotics, phages are increasingly considered as alternatives to target drug-resistant bacterial pathogens. Over the years, several phase I clinical trials have demonstrated the safety of phages and various case studies show how phage therapy can be both effective and lifesaving in cases where no other treatments are available (Schooley et al., 2017; Dedrick et al., 2019; Eskenazi et al., 2022).
However, phage therapy has been difficult to scale to the many thousands or more that could benefit from it (Ireland, 2024). The three major reasons for this are:
- Most phages engage in very specific interactions with their bacterial hosts. This makes it difficult to find the one or few matching phages against a pathogen of interest.
- Phage manufacturing and logistics are difficult. Producing a phage in a purified solution at a sufficient concentration can be tricky. And if the phage is needed at another location or country, that makes things even trickier.
- Traditional legislation is not a good fit for phage therapeutics. Phages are biological entities that are unlike the typical drugs we develop. This means that, in most countries, today phages can only be used as a very last resort as compassionate care. Different countries can also have different specific rules.
We can use artificial intelligence to find matching phages a lot faster
In our work, we are tackling the first bottleneck. Most phages are quite specific, and this is problematic because it necessitates a specific search for one or more matching phages against a particular bacterial pathogen. In the lab, this can become a time and labor intensive process, and it does not scale well to screening large collections of 100’s or even 1000’s of phages. This led us to the question: can we develop computational tools that can screen phages in silico in a way that is practically relevant? In particular, we want to make predictions at the most specific level of phage-host interactions: the bacterial strain level.
Now, to train an AI model, you need a sufficient amount of data to do so. For most bacterial species this is still a bottleneck. We have been fortunate to get in contact with the EnBiVir Lab in Valencia, which had characterized around 10,000 phage-host interactions for Klebsiella bacteria, together with their genomic sequences. This provided us with a great starting point to develop predictive models.
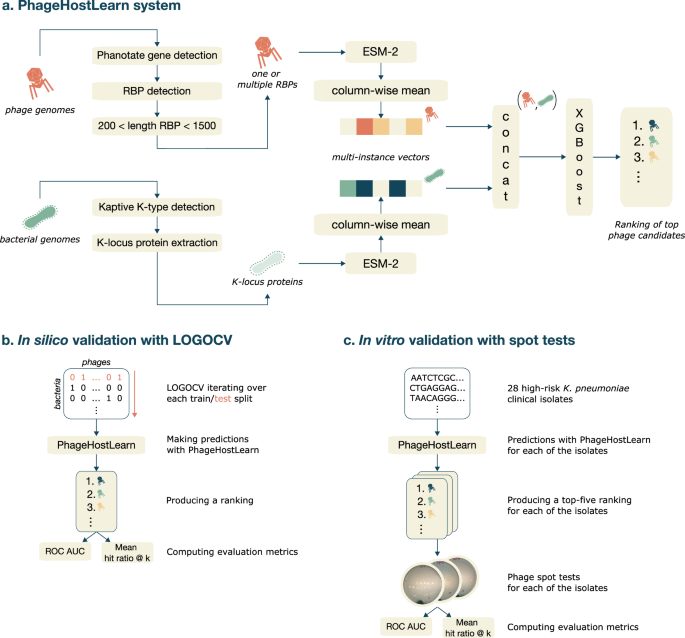
So that's what we did. We have developed PhageHostLearn, a model that can predict Klebsiella phage-host interactions at the strain level, and which provides the predictions in a very practical output format as a ranking of phages to test against a particular bacterium. Specifically, we have focused on the very first step of an interaction between a phage and a Klebsiella bacterium: when the phage touches the surface of the bacterial host and interacts with proteins and other surface receptors. For Klebsiella phage-host interactions, this if often the most important step in the phage's infection cycle. Correspondingly, we have trained our model by giving it the specific proteins involved at both the bacterial side (the CPS surface receptors) and the phage side (the receptor-binding proteins or RBPs).
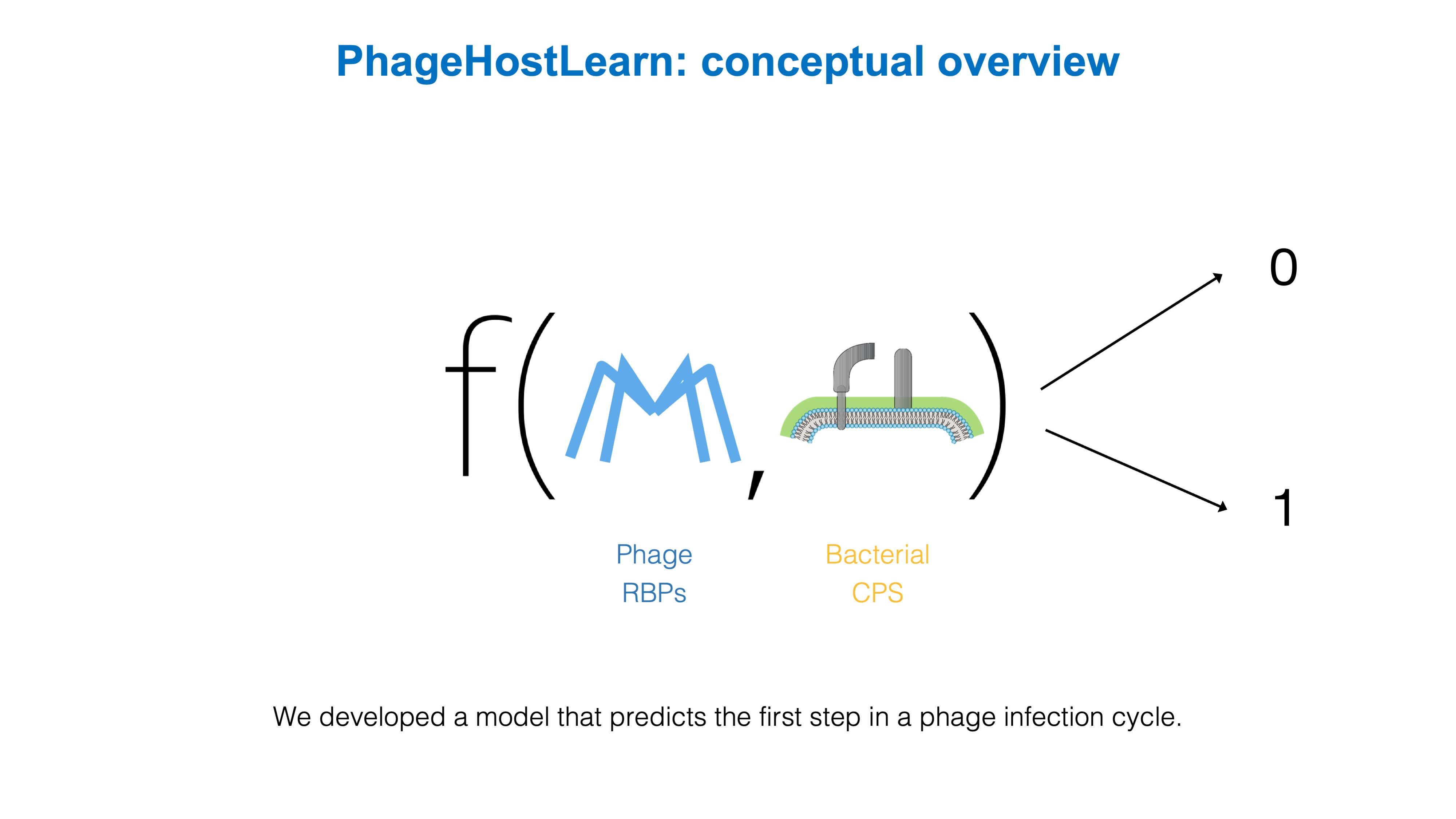
We show that our model is successful in predicting interactions with this information, and have put it to the test by letting it predict interactions for high-risk Klebsiella pathogens that are currently circulating in Spain and which the model has not seen before. In addition, we measure a practical and easy-to-understand metric: the average probability of finding at least one matching phage in a top k of suggested phages by the model, known as the hit ratio @ k. For example, with our model we expect to find at least one 'hit' in the top-10 in around 65% to 84% of the cases on average. We think this is a very useful metric because it can provide researchers and clinicians with a very practical answer to the question: how many phages will I have to test to find one that works?
Where to go from here?
Over the last years, the progress in both the phage research community and the AI research community has been nothing short of impressive. To us, it is increasingly clear that AI methods can be incredibly useful to help solve previously intractable problems in biology and medicine. We see our work as a specific case study and positive evidence of that. Nevertheless, there is a lot of progress yet to be made. Related to our work specifically, it would be very useful to have models able to predict interactions for various important bacterial species. Large and diverse sets of phage-host interaction data are crucial to enable this (we would even want current Klebsiella datasets to be an order of magnitude bigger). In turn, we are convinced that such models could meaningfully contribute to more effective phage therapeutics and diagnostics, to help tackle the increasingly big problem of antimicrobial resistance.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in