Finding the keys to exit the host cell
Published in Microbiology

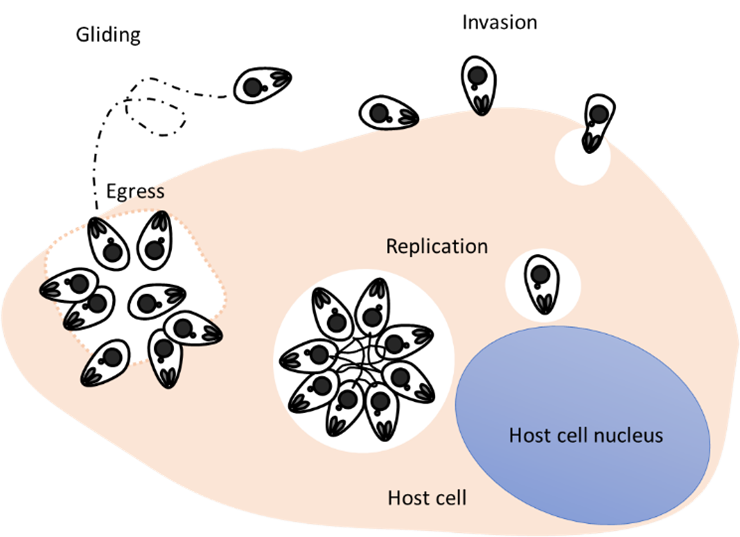
In this study, we were determined to explore the possibility of finding genes important for different aspects of the Toxoplasma gondii life cycle, with a special interest on changes in actin dynamics. Parasite actin is important not only for the motility of these obligate intracellular parasites and their invasion of the host cell, but also for communication in between individuals within their parasitophorous vacuole, and during their egress1.
Establishing a new technology for phenotypic screens. At this point everyone knows that CRISPR/Cas9 has revolutionised genetic modifications in various organisms, where it has been successfully applied. In our field, Cas9 has been employed not only to target individual genes, but also to screen all annotated genes in this parasite to discover genes vital during the asexual stage2. However, we were only able to infer the importance of a certain gene based on the fact that its absence would kill the parasite. While this is an important information, the function during the parasite’s life cycle remained unknown without additional, time-consuming experiments. With this in mind, we decided to adapt a conditional Cas9-system so that only in the presence of an inducer a gene of interest (GOI) will be disrupted allowing phenotypic characterisations. We therefore introduced a Cas9 split in two halves (splitCas9)3 that will only be active after the addition of rapamycin.
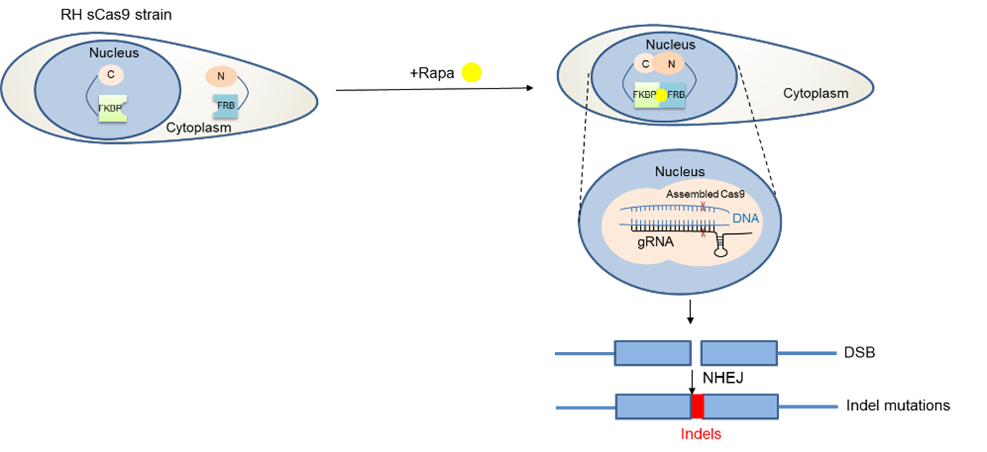
After optimisation of this powerful technology, we decided to performed a phenotypic screen on 320 selected, hypothetical and essential genes that are only present in T. gondii and related species (apicomplexan parasites). We inserted a library of vectors containing sequences (gRNAs) targeting those genes into an indicator parasite strain that expressed splitCas9 and fluorescent markers for actin, allowing us to test for differences in actin-dynamics in live parasites upon disruption of a gene of interest. These parasites were subsequently sorted into 96 well plates to isolate clonal lines. After induction with rapamycin, we performed imaging and compared the morphology of the respective parasites to identify differences in actin dynamics and host cell egress.

After the selection of interesting phenotypes, we identified the respective gRNAs in 267 clones by sequencing. We identified 99 unique gRNAs from which we selected 42 for further exploration. After validation of the results and tagging of 16 of the most promising candidates, 2 of them stood out over the rest: CGP (conoid gliding protein) and SLF (signal linking factor).
Two apical proteins essential for parasite exit from the cell. CGP was annotated as a hypothetical protein with unknown function. Here we localised CGP to the conoid, a tubulin barrel-like structure that extends after egress, during motility and invasion. By applying a conditional knock out strategy (cKO) based on a rapamycin-induced dimerisable Cre-recombinase(DiCre)5, we validated the phenotype observed in our screen and found that parasites lacking CGP are incapable of host cell egress, motility and invasion. The second candidate was SLF, previously identified in a pull-down as an interacting protein of a signalling complex that is involved in egress6. However, in this study SLF has been reported as dispensable, since knockdown did not result in parasite death6. Though using a different conditional system that allows complete removal of a gene of interest, we found that SLF is crucial for the parasite invasion, motility and egress. Moreover, deletion of SLF led to disassembly of the remaining components of the signalling platform.
Interestingly, closer analysis of phenotypes caused by disruption of SLF and CGP demonstrated that these proteins are crucial for independent steps during host cell egress, since different and distinctive behaviours with respect to the changes in actin dynamics and parasitophorous vacuole integrity were observed.
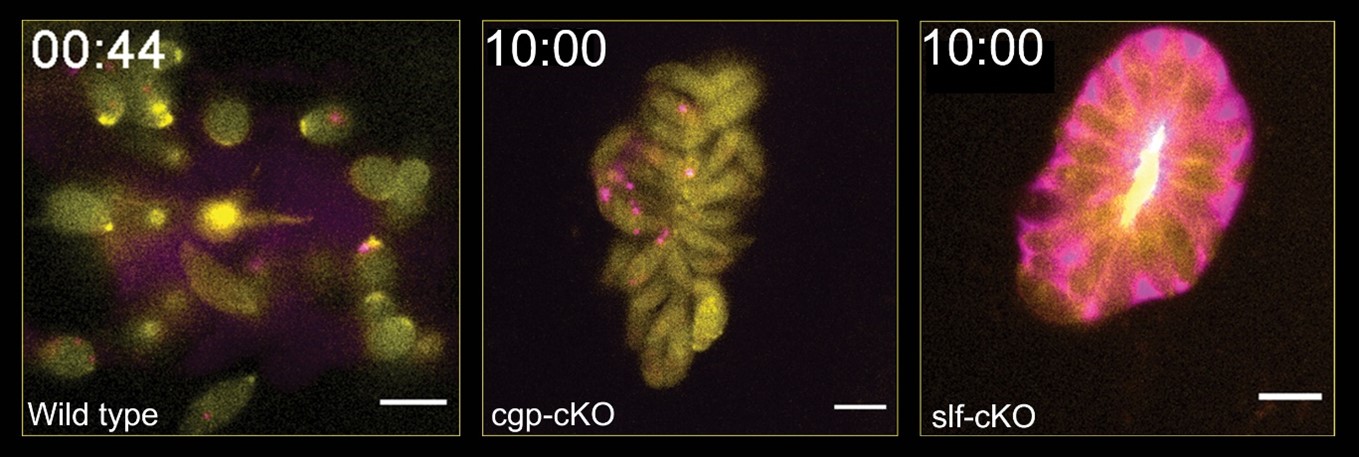
While deletion of slf led to an early block in host cell egress, since these parasites were unable to dissolve the intravacuolar actin-network or the parasitophorous vacuole, deletion of cgp resulted in parasites capable of dissolving both. However, parasites remained inmotile therefore incapable to leave the host cell.
At the moment, we are continuing to study the function of these proteins in more detail and to identify their interaction partners to gain mechanistic insights into both processes that are crucial for host cell egress.
This exciting project has led us to new interesting avenues of research. For example, we modified our indicator splitCas9 strain to look for genes involved in secretion of important virulent factors, mitochondrial stability, as well as cytoskeleton assembly and maintenance.
1 Periz, J. et al. Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. eLife 6, doi:10.7554/eLife.24119 (2017).
2 Sidik, S. M. et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 166, 1423-1435.e1412, doi:10.1016/j.cell.2016.08.019 (2016).
3 Zetsche, B., Volz, S. E. & Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology 33, 139-142, doi:10.1038/nbt.3149 (2015).
4 Li, W., Grech, J., Stortz, J.F. et al. A splitCas9 phenotypic screen in Toxoplasma gondii identifies proteins involved in host cell egress and invasion. Nat Microbiol (2022). https://doi.org/10.1038/s41564-022-01114-y
5 Andenmatten, N. et al. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nature methods 10, 125-127, doi:10.1038/nmeth.2301 (2013).
6 Bisio, H., Lunghi, M., Brochet, M. & Soldati-Favre, D. Phosphatidic acid governs natural egress in Toxoplasma gondii via a guanylate cyclase receptor platform. Nature microbiology 4, 420-428, doi:10.1038/s41564-018-0339-8 (2019).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in