Flexibility meets quality and scale
Published in Protocols & Methods and Cell & Molecular Biology
Similar to single-cell RNA sequencing, single-cell mass spectrometry proteomics met a substantial challenge with sample preparation. Some of the challenges are shared with other single-cell methods, such as maximizing sample delivery to the instruments and scaling up to thousands of cells. Others are more specific to mass spectrometry, such as minimizing all sorts of contaminants that interfere with mass spectrometry measurements and the need to support diverse multiplexing workflows. Finding solutions to all of these challenges was a major bottleneck for the burgeoning field of single-cell mass spectrometry proteomics.
The physicochemical diversity of proteins poses challenges to efficiently delivering proteins to instruments for analysis. Different amino acid sequences can adsorb to various surfaces, thus resulting in losses, reduced sensitivity and even biases in protein sequence coverage. These problems may be especially severe with small samples, such as single cells and were among the challenges that we described in a 2018 perspective highlighting challenges and opportunities for single-cell proteomics. To minimize these losses, our nPOP protocol (described by Leduc et al., 2022, 2024) uses fluorocarbon surfaces to repel both hydrophilic and hydrophobic peptides. Indeed, measurements suggest that peptides from single cells are delivered to mass spectrometry detectors with about 95% of the efficiency for delivering peptides from samples composed of thousands of cells.

Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP, Nat Protoc (2024). 10.1038/s41596-024-01033-8
Our preferred solution for the challenges of single-cell proteomics was to prepare single cells on flat glass slides that maximize the freedom for droplet deposition, just as blank canvas opens space for creativity. At the same time such open experimental design allows for increasing the density of droplets for sample preparation and therefore the number of single cells that can be processed and prepared in parallel. This simple solution was everything but simple to implement. Fortunately, Andrew Leduc embraced those simple ideas and found elegant and resourceful ways to resolve the numerous challenges along the way of their practical implementation. For example, he found that DMSO can be used both to lyse single cells and as a solvent for labeling reactions to increase the robustness of the protocol. He rigorously benchmarked every step and iteratively found better solutions for problematic steps. Solving problems and optimizing numerous parameters step by step, Andrew developed a robust protocol that not only improved the quality of sample preparation but also increased its scale and flexibility. Going a step further, Andrew made the protocol accessible, developed a computational package (QuantQC) for processing data, and helped others adopt it. We hope you enjoy its flexibility and scalability as much as we do and use its high quality data for exciting discoveries.
Leduc, A., Khoury, L., Cantlon, J. et al. Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP. Nat Protoc (2024). doi: 10.1038/s41596-024-01033-8
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcement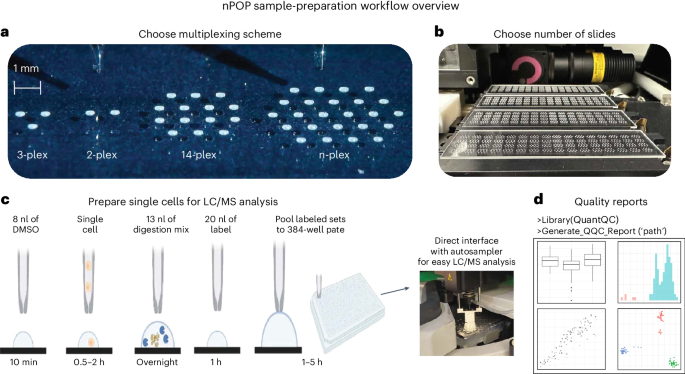

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in