FOXQ1 utilizes the KMT2A/MLL complex as a transcriptional co-activator of EMT in breast cancer
Published in Cancer
FOXQ1 recruits the MLL complex to activate transcription of EMT and promote breast cancer metastasis
Epithelial-to-mesenchymal transition (EMT) has been proposed as a frequent mechanism contributing to drug resistance, recurrence, local invasion, and distant metastasis in many carcinoma types, including breast cancer. This process involves the activation of mesenchymal-associated genes and the suppression of epithelial genes. While numerous microenvironmental cues and signaling pathways have been identified as capable of inducing an EMT in cancer, the crux of EMT regulation is thought to rely on a limited number of EMT transcription factors (TFs).
Studies conducted in the Wu lab at the Karmanos Cancer Institute previously identified the Forkhead box transcription factor, FOXQ1, as an exemplar EMT-TF in the aggressive, triple-negative subtype of breast cancer (TNBC) (1). In the decade since this work was published, the role of FOXQ1 as an EMT-TF has been affirmed in models spanning 13 carcinoma types. Further, FOXQ1 expression is prognostic of worse overall survival in patients across six cancer types, including breast (2).
The molecular mechanisms responsible for the regulation of FOXQ1 transcriptional activity have not been characterized. Therefore, this work set out to uncover the epigenetic mechanisms employed by FOXQ1 to regulate the transcriptional state of downstream EMT effectors.
Using a proteomics-based approach, we found that FOXQ1 can complex with several subunits of the KMT2/MLL histone methyltransferase family. The KMT2/MLL complex is well characterized to facilitate histone-3, lysine-4 trimethylation (H3K4me3) within the chromatin regulatory elements of actively transcribed genes. KMT2/MLL family has low intrinsic enzymatic activity and requires the binding of a three-subunit core complex (containing RbBP5, ASH2L, and WDR5) to achieve sufficient activity for H3K4 trimethylation (3). Using multiple biochemical approaches, FOXQ1 was found to bind a complex containing the KMT2A/MLL1 histone methyltransferase by directly binding to the RbBP5 subunit.
Through ChIP and RNA- sequencing, we found abundant chromatin co-localization of FOXQ1 and RbBP5 within the promoter regions of genes with critical EMT functions including several critical TFs: FOXC2, TWIST1, ZEB1, and SIX2. Overall, 92% of FOXQ1-bound active promoters harbored coincident RbBP5 binding sites. Mechanistically, we found that both RbBP5 and MLL1 were required for the downstream transcriptional activation of FOXQ1 target genes, however, FOXQ1 promoter recognition was found to precede RbBP5/MLL1 chromatin binding. Altogether demonstrating that FOXQ1 recruits RbBP5 to the target promoter regions to facilitate MLL1 complex assembly, H3K4me3 deposition, and initiation of transcriptional activation.
We also identified several amino acids within the Forkhead box domain (FHD) of FOXQ1 that are critical for RbBP5 binding without interrupting the binding of FOXQ1 to its downstream promoter targets. Therefore, our study identified a FOX FHD harboring a second essential function as a transactivation domain. Ectopic expression of FOXQ1 mutants, which were defective for RbBP5-binding, were no longer capable of inducing EMT in breast epithelial cells suggesting that the KMT2A/MLL1 complex is a necessary co-factor for FOXQ1 to function as an EMT-TF. Further, targeting the FOXQ1-MLL axis was able to reverse the EMT phenotype and lead to a decrease in spontaneous metastatic progression in vivo.
Our data support a two-step model in which FOXQ1 recognizes and binds to specific target promoters (a step independent of the MLL core complex) but has minimal effect on the transcriptional activation. FOXQ1 then recruits RbBP5, which facilitates MLL complex assembly and deposition of H3K4 trimethylation (see image below). Overall, the FOXQ1-MLL axis may be critical for understanding the epigenetic regulation of EMT, and therapeutic strategies to disrupt this epigenetic axis warrant further exploration.
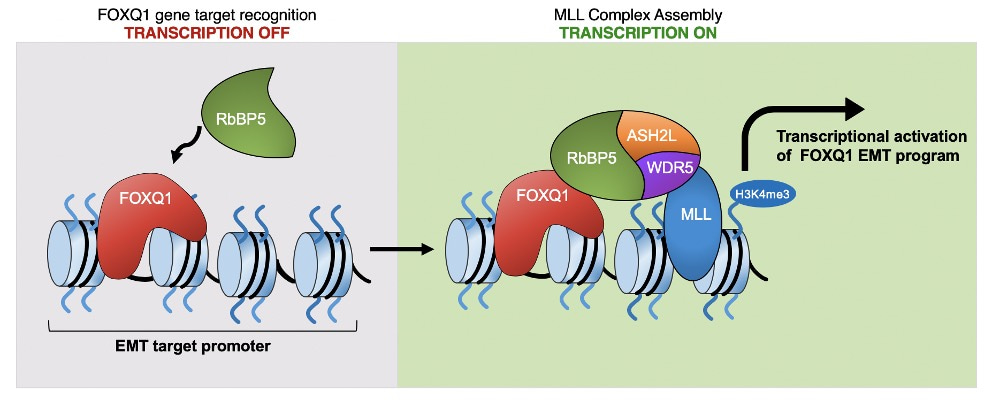
A proposed model illustrates that FOXQ1 directly recruits RbBP5 to the promoters of EMT target genes to facilitate MLL complex assembly, H3K4me3, and transcriptional activation.
References:
1. Zhang H, et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer research 71, 1292-1301 (2011).
2. Cui X, et al. Prognostic value of FOXQ1 in patients with malignant solid tumors: a meta-analysis. OncoTargets and therapy 10, 1777-1781 (2017).
3. Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature structural & molecular biology 13, 713-719 (2006).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in