From Uganda to the UK: Lessons in Viral Sequencing
Published in Microbiology

What began as a straightforward knowledge exchange between two MRC Units, quickly expanded into something far greater—a deep and enduring collaboration that spanned continents, multiple research projects, three large collaborative grants and a significant investment of time and effort. Both MRC Units had been awarded Illumina MiSeq next generation sequencing platforms as an investment into virus sequencing by the MRC. As we engaged with our Ugandan colleagues, our partnership grew into a project resulting in the co-supervision of six PhD students - three from each country (Stella Atim, Martin Mayanja, Prossy Namuwulya, James Shepherd, Leah Owen, Saoirse Healy), fostering joint positions for talented postdoctoral scientists—including Hannah Jerome, Chris Davis and later Stella Atim and Shirin Ashraf—and undertaking far more work than we had originally envisioned. The vision of Professor Pontiano Kaleebu and Dr Josephine Bwogi at UVRI and more recently Professor Moffat Nyirenda has led to the implementation of sequencing at this centre, working both with CDC and the MRC. This has led the way to the discovery of new viruses, something that the Uganda Virus Research Institute has been renowned for since it's inception just over 100 years ago, although previously with more traditional virus discovery methods. What started as a sequencing workshop turned into a transformative learning experience for all involved. We spent countless hours refining our sequencing methodologies, improving both our wet lab techniques and bioinformatics pipelines, ensuring that our analyses were not only accurate but also reproducible at scale.
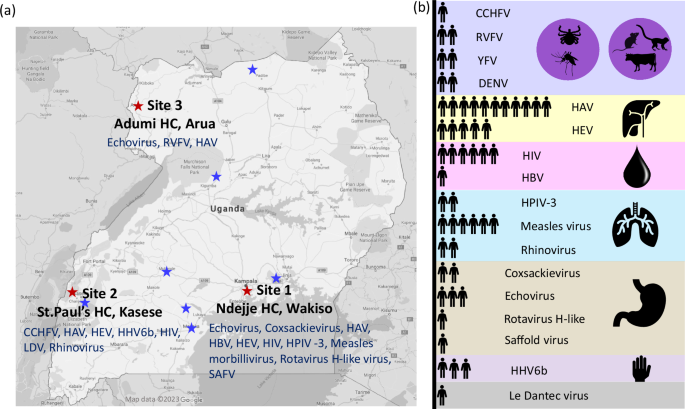
Our findings indicated that viruses that cause hemorrhagic fever are circulating under the radar in Uganda, and led on to surveys that suggest that around 1 in 4 people in some farming communities in Uganda have been exposed to Crimean-Congo hemorrhagic fever virus (CCHFV) and 9 in 10 cattle. We have now regularly detected and sequenced the virus and several related viruses (including Dugbe virus and NSDV) in ticks (mostly in Rhipicephalus ticks). This high rate of exposure indicates a substantial health burden that is only partially documented to date and has led on to a study funded by NIH and BBSRC to investigate possible cross-reactivity to different orthonairoviruses in Uganda.
One particularly valuable lesson from this work emerged in the supplementary data of our study. While analysing samples, we detected what appeared to be a novel virus. However, after extensive investigation—including repeat extractions, PCR assays, and multiple rounds of sequencing—it became evident that this was most likely a contaminant originating from the blood collection tubes rather than a genuine new pathogen. The absence of seroconversion in patients reinforced this conclusion. This experience underscored the importance of rigorous quality control in sequencing studies and the necessity of critically evaluating unexpected findings.
While this discovery took several years to resolve, the expertise we gained through these meticulous validation efforts has been invaluable. Today, we are able to conduct sequencing analyses with far greater speed and precision. For example, in our recent publication in Nature, we identified adeno-associated virus 2 (AAV2) in children with hepatitis in the UK in real time during the peak of the outbreak. The skills honed through our work in Uganda directly contributed to this work, demonstrating how foundational research can lay the groundwork for future discoveries.
Scientific progress is rarely linear. It often involves detours, unforeseen challenges, and hard-earned lessons. Our collaboration with UVRI and Makerere University has not only strengthened sequencing capacity in Uganda but has also profoundly shaped our own research capabilities and deepened our friendships and understanding across different cultures. This partnership exemplifies the power of international scientific exchange—where knowledge shared leads to discoveries that benefit global public health. And as we continue our work, we carry forward the lessons from the AFI study, ensuring that our sequencing efforts remain robust, reliable, and impactful.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in