Generating sterile Anopheles mosquitoes to combat malaria transmission
Published in Protocols & Methods, Genetics & Genomics, and Zoology & Veterinary Science

Malaria remains a deadly disease, claiming hundreds of thousands of lives worldwide every year. The primary vectors of malaria are Anopheles mosquitoes, including Anopheles gambiae and Anopheles stephensi. Controlling mosquito populations is crucial for stopping malaria transmission. Gene drive technology represents a promising alternative to traditional chemical-based control methods, offering a species-specific and eco-friendly approach. However, key challenges in developing gene drives include resistance formation due to end-joining repair after Cas9/gRNA cleavage and the fitness costs associated with drive heterozygotes.
One of the most successful gene drive designs to date targets the doublesex (dsx) gene in Anopheles gambiae, as reported by Kyrou et al. (2018). The dsx gene plays a crucial role in sex differentiation, with distinct isoforms expressed in males and females. Disrupting the female-specific isoform results in female sterility. Ideally, only homozygous females exhibit sterility due to a male-like intersex phenotype, while male homozygous and heterozygous mosquitoes of both sexes remain fertile. This makes it a suitable target for a suppression gene drive aimed at reducing the number of fertile females and ultimately eliminating the population.
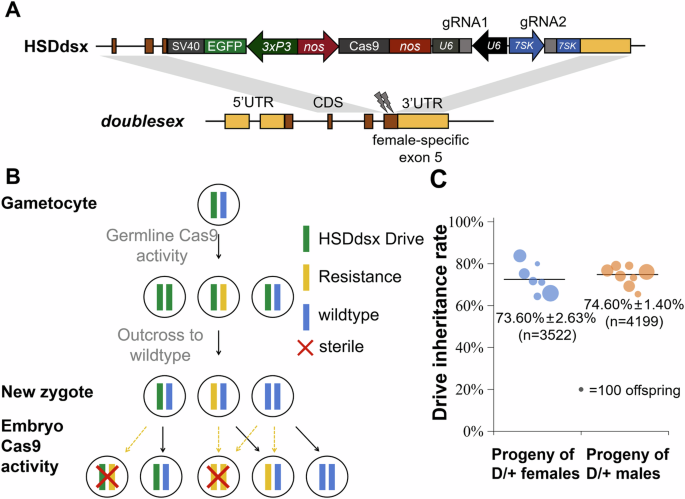
The specific target site for this gene drive was strategically placed at the intron 4/exon 5 boundary. This design minimized the risk of functional resistance because mutations at this site were likely to disrupt dsx function, rendering resistance alleles nonfunctional (not expressing a correct female dsx product). However, since only a single gRNA was used in the original dsx suppression drive, functional resistance may still arise in larger populations. Another limitation was the fitness cost in female drive heterozygotes due to unintended Cas9 activity in somatic cells, potentially reducing the effectiveness of population suppression.
Based on the previous modeling research by S. Champer et al. (2022), we hypothesized that using the nanos promoter for Cas9 expression could reduce fitness costs compared to the zpg promoter used in the original A. gambiae study. Given the close evolutionary relationship between A. gambiae and A. stephensi, we expected similar outcomes in A. stephensi. To further enhance the robustness of the drive, we designed a system with multiplexed gRNAs, aiming to reduce functional resistance allele formation.
To assess drive efficiency, we first crossed drive heterozygotes with wild-type mosquitoes. Phenotyping of the offspring revealed moderate drive conversion rates in both male and female germlines. However, the intercross of drive heterozygotes yielded unexpected results—an unusually high proportion of intersex mosquitoes. In a typical mosquito population, the expected sex ratio is approximately 50:50. The numbers of morphologically male and female mosquitoes were similar, even though we expected more males (because female homozygotes would be male-like intersex). Genotyping revealed that both genetically male and female homozygous mosquitoes exhibited intersex phenotypes and were sterile. Despite conducting several molecular tests, the exact cause of sterility in male homozygotes remains unclear.
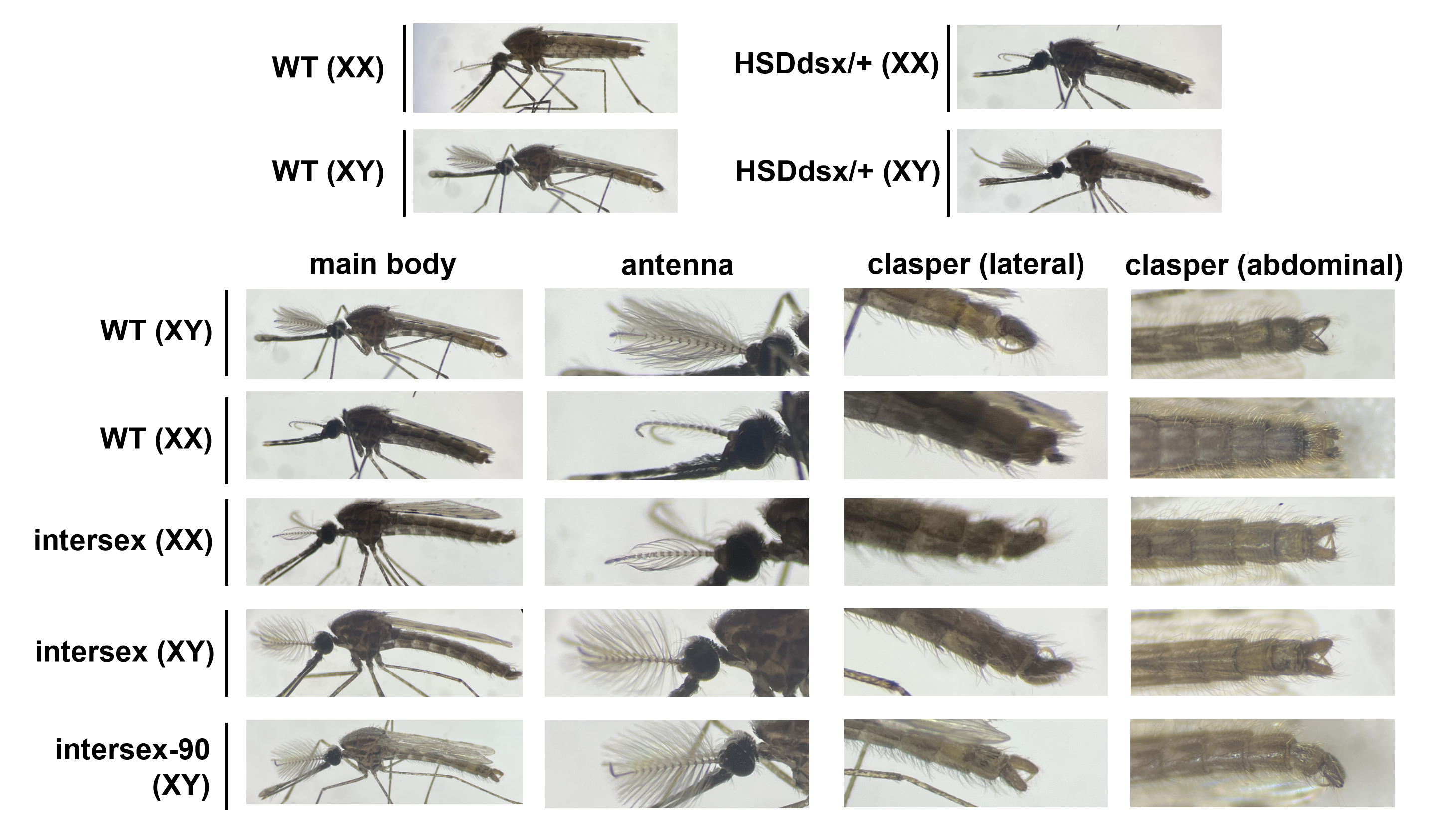
The morphology analysis of our suppression drive line. Heterozygous drive males and females showed normal morphology like wild-type, while various intersex phenotypes were observed in homozygotes. XX and XY indicate genetic females and males, respectively, and all the intersex mosquitoes were drive homozygotes, as confirmed by genotyping.
This outcome potentially differs from the original A. gambiae study, where only female homozygotes were reported as intersex. Also, even though it supported moderate drive conversion, the nanos promoter for Cas9 yielded lower efficiency in A. stephensi. Our findings highlight the species-specific nature of gene drive targets—what works effectively in one species may not be a universal solution for others. Computational modeling indicated that our suppression drive would only be successful if male homozygotes remained fertile and if competing species or predators also contributed to population decline. These results underscore the necessity of species-specific assessments when designing suppression drives.
To evaluate resistance formation, we performed amplicon sequencing on mosquitoes derived from intercrossed drive heterozygotes. A very low level of resistance was detected, with most mutations appearing to be somatic and therefore unlikely to be transmitted to the next generation. Additionally, the detected resistance mutations were not in-frame, meaning they did not preserve functional dsx protein. Our design incorporated two gRNA target sites, and the frequency of both sites mutating simultaneously was even lower than that of a single-site mutation. This suggests that our multiplexed gRNA strategy likely effectively mitigated functional resistance allele formation.
To enhance drive efficiency, we combined our suppression drive with a vasa-Cas9 line, which boosted drive conversion rates to 100% in males while rendering female drive heterozygotes sterile. Although the spread ability of this drive was reduced due to somatic cleavage of dsx by vasa-Cas9 in these females, this combination gene drive system could be used as a self-limiting strategy for population suppression. This kind of gene drive is more controllable than the original homing drive design. Modeling of a similar combination system demonstrated successful population suppression with a lower male release requirements than other self-limiting approaches such as Sterile Insect Technique (SIT) and female-specific RIDL (fsRIDL), both of which have been deployed in the field. This suggests that our combined system could provide a viable alternative for vector control.
Our findings offer novel insights for improving suppression drives and developing release candidates for the field deployment, while emphasizing the necessity of species-specific assessments when selecting target genes for population control. Despite remaining challenges, our work contributes to the advancement of gene drive technology as an efficient and eco-friendly solution for malaria vector control.
References:
Champer, S.E., Kim, I.K., Clark, A.G., Messer, P.W., Champer, J., 2022. Anopheles homing suppression drive candidates exhibit unexpected performance differences in simulations with spatial structure. Elife 11, e79121. https://doi.org/10.7554/eLife.79121
Kyrou, K., Hammond, A.M., Galizi, R., Kranjc, N., Burt, A., Beaghton, A.K., Nolan, T., Crisanti, A., 2018. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol 36, 1062–1066. https://doi.org/10.1038/nbt.4245
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Step by step towards a more malaria-resilient world. Well done! I have included other publications from this group in my living literature review on gene drive, Monitoring Gene Drives.