How bacterial regulation of p53 shapes tumorigenesis
Published in Cancer
The human microbiome has emerged in the recent years as an important modulator of tumor initiation and progression1, and its alteration has been recently added to the hallmarks of cancer2. Several oncogenic bacteria, such as Helicobacter pylori or genotoxin-producing E. coli, have been described as potential cancer drivers. Other species have also been associated with increased cancer risk, such as Fusobacterium nucleatum or Klebsiella pneumoniae. Overall, the bacterial molecular mechanisms promoting cancer and how they affect the host pathways remain poorly characterized.
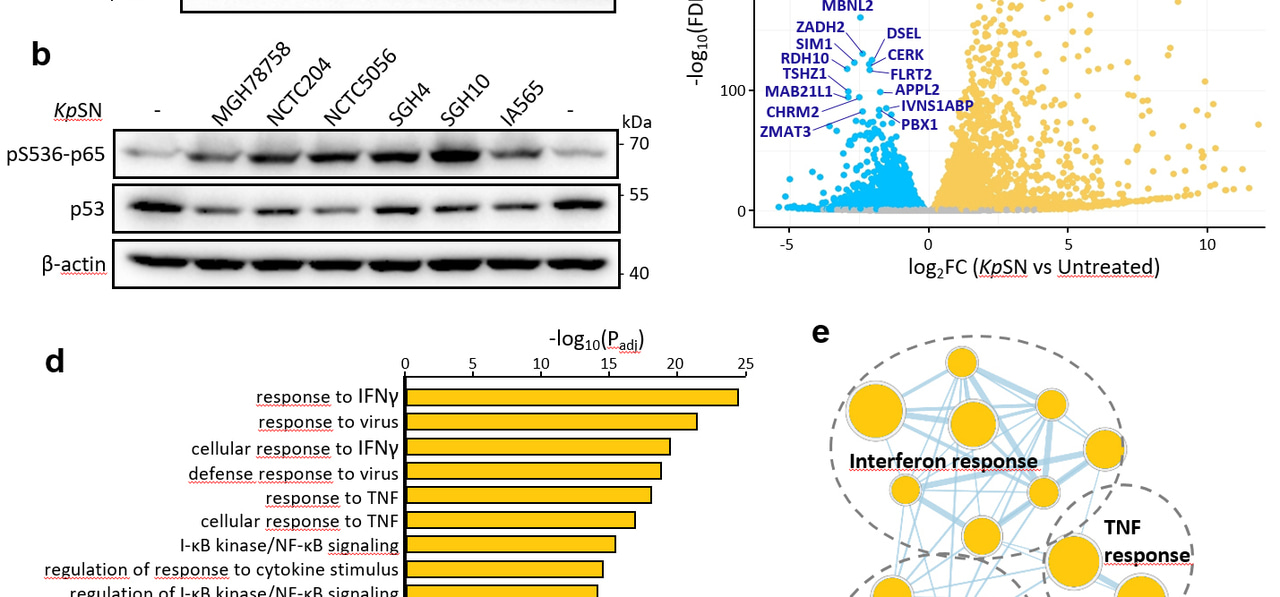
Using in-vitro and ex-vivo infections of human cells by Klebsiella peumoniae, we have found that lipopolysaccharides (LPS) associated with K. pneumoniae outer membrane vesicles (OMVs) were strongly inhibiting p53 protein level and activity. Inactivation of p53 is a major step toward tumorigenesis due to its transcriptional regulation of tumor suppressive programs. Upon chemical induction of DNA damage (by doxorubicin) or oncogenic stress induced by Ras, we found that p53 was not able to trigger its normal transcriptional response or to induce cellular senescence when exposed to K. pneumoniae OMVs. We identified that p53 was repressed not only by K. pneumoniae OMVs but also by OMVs from other Enterobacteria, but not by commensal Gram negative bacteria such as non-toxigenic Bacteroides fragilis, which highlights the role of bacterial dysbiosis during cancer initiation.
Exploring the mechanism of p53 inhibition, we found that p53 was repressed through the classical LPS-TLR4-NFκB pathway, which has already been proposed to antagonize p53 during inflammation3. We hypothesized that p53 inhibition by bacterial signaling could alleviate the selection pressure for p53 mutation during carcinogenesis. Using patient data from The Cancer Genome Atlas (TCGA), we found that p53 mutation rate is inversely correlated with the expression of TLR4 in early grade colon cancers, but not in other types of cancer with low abundance of gram negative bacteria (such as breast cancers). Interestingly, differences in the microbiota composition between wild type and mutant p53 tumors have already been described4. Altogether, these data suggest that bacterial signaling can act as an evolutionary driver during cancer and shape tumor evolution.
Importantly, we have discovered that p53 was inhibited through the destabilization of its mRNA but not at the level of protein stability. p53 has been so extensively characterized to be regulated by MDM2-ubiquitin-dependent proteasomal degradation that the regulation of its mRNA is often overlooked. We were very surprised that inhibition of p53 by K. pneumoniae OMVs could not be rescued by proteasome inhibitors or MDM2 inhibitor Nutlin, and that overall the protein half-life of p53 was not affected. We identified that LPS triggers an NFκB-dependent inhibition of the gene ZMAT3, which encodes a RNA binding protein normally stabilizing p53 mRNA. Importantly, ZMAT3 is also a target gene of p53, forming a feedback loop which amplifies p53 inhibition.
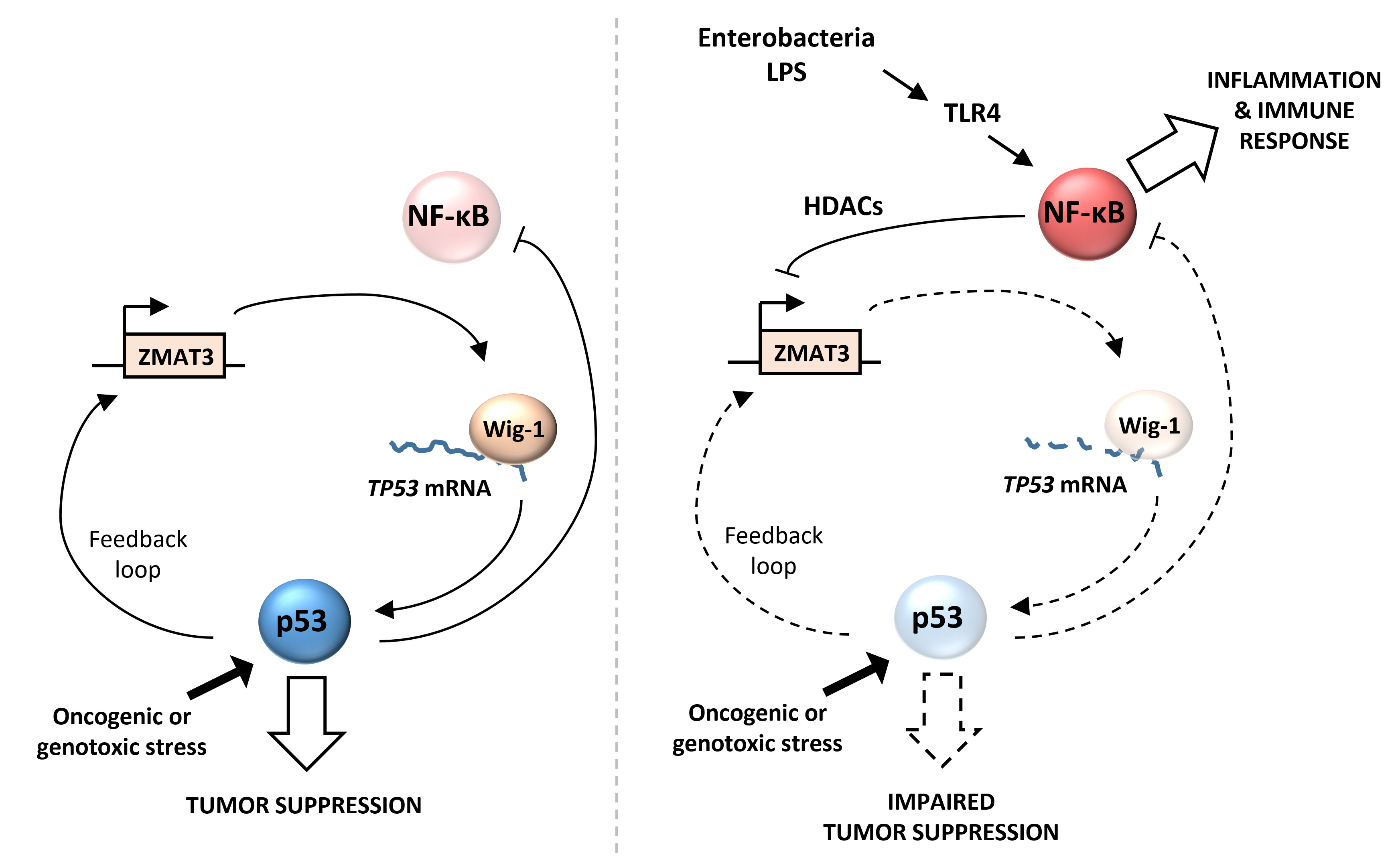
Overall, our results suggest that ZMAT3-p53 feedback loop could act as a molecular switch between NFκB and p53 signaling during inflammation. As p53 and NFκB are two antagonistic pathways, inhibition of p53 could be an important step to allow a proper NFκB activation and inflammatory response to bacterial infections. On the other hand, this inhibition of p53 could create a pro-tumorigenic environment, especially in the context of chronic inflammation.
References
- Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, (2021).
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 12, 31–46 (2022).
- Gudkov, A. V. & Komarova, E. A. p53 and the Carcinogenicity of Chronic Inflammation. Cold Spring Harb. Perspect. Med. 6, (2016).
- Greathouse, K. L. et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123 (2018).
Follow the Topic
-
Oncogene

This journal aims to make substantial advances in our knowledge of processes that contribute to cancer by publishing outstanding research.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in