How did the orchid originate and evolve?
Published in Ecology & Evolution

Worldwide, there are nearly 30,000 orchid species. They show unique flower morphologies (petals with a specialized labellum, stamens and pistil fused into a gynostemium, a pollinium, and tiny seeds without endosperm); grow on the ground, trees, or rocks with an extraordinary diversity in lifestyle; and have successfully colonized almost every place on Earth1-3. The origin, evolution, and diversification of orchids has always been a mystery4. We carried out whole-genome sequencing of Apostasia shenzhenica (Fig. 1) and whole-genome re-sequencing of Phalaenopsis equestris (Fig. 2) and Dendrobium catenatum (Fig. 3), and combined these data with those from the transcriptomes of other orchid species and non-orchid species and from gene function analyses. The study reveals the molecular mechanisms and the evolutionary paths of orchid flower development, lifestyle, and diversification, and also successfully unveils the mystery of orchid evolution5.

Figure 1 An A. shenzhenica plant is flowering in the wild of Shenzhen, China.
Genome sequencing and assembly
We used a new sequencing strategy based on second-generation sequencing combined with 10X genomics and third-generation sequencing technologies. This sequencing strategy facilitated high quality assembly of the genomes of A. shenzhenica (scaffold N50 3.029 Mb, contig N50 80.1 Kb), P. equestris (scaffold N50 1.217 Mb, contig N50 45.79 Kb), and D. catenatum (scaffold N50 1.055 Mb, contig N50 51.7 Kb). The genome size of A. shenzhenica is estimated to be 471.0 Mb. We annotated 21,841 protein-coding genes, which is more than 7,000 genes less than in P. equestris (29,545) and D. catenatum (29,257). Furthermore, we collected various tissues of representative orchid species from five subfamilies, and obtained large amounts of transcriptome data for use in gene functional analysis and validation.

Figure 2 A flowering plant of P. equestris of the subfamily Epidendroideae. Its whole genome has been re-sequenced for this study.
Whole-genome duplication (WGD) and the origin of orchids
The comparative genomics analysis suggested that the ancestor of all extant orchids underwent a WGD event, which occurred before the third biological extinction event (66 Mya)6(Fig. 4). After the WGD event, the ancestral orchid added one further set of genes, and successfully survived extinction. After the biological extinction, there was widespread destruction of ecological systems. However, orchids rapidly diverged and evolved into five subfamilies through gene family expansions and contractions, producing high diversity and adapting rapidly to the new ecosystems. Consequently, almost all extant orchids in the world are the offspring of orchids that survived through the period of dinosaur extinction.

Figure 3 The plants of Tie Pi Shi Hu D. catenatum (Epidendroideae) were growing on the tree. The whole genome of this species has been re-sequenced for this study.
Origin and evolution of orchid morphology
By comparing 12 plant species, we identified 474 orchid-specific gene families, permitting the reconstruction of an ancestral orchid gene toolkit, from which we were able to gain new insights into the evolutionary history of new gene families and gene family expansions and contractions, particularly for the “life” and “death” historical traces of MADS-box genes, which control orchid flower development. These analyses revealed the molecular mechanisms underlying the development of the labellum, gynostemium, pollinium, and seeds without endosperm, and the evolution of epiphytic and terrestrial lifestyles.
We identified 36 putative functional MADS-box genes that regulate orchid flower organ development. Previous studies have shown that development of the labellum and gynostemium in orchids is regulated by the B-AP3 and E classes of these genes7. These two classes of genes existed in the common ancestor of all extant orchids, which underwent WGD. The loss of B-AP3 and E classes in A. shenzhenica has resulted in loss of the labellum and an incomplete gynostemium. After the WGD in the orchid ancestor, the duplicated B-AP3 genes regulated the development of petals with a differentiated labellum, producing a bilaterally symmetrical flower structure. Flowers with bilateral symmetry improve the selection efficiency of specific pollinators and accuracy of pollen falling position (genes involved in pollinium formation also improve the precision of pollen transfer). This consequently ensured their reproductive success and also promoted the production of new orchid species and the development of high diversity, which is also a reason for the existence of more flowers with bilateral symmetry than with radial symmetry. These findings have overturned previous perceptions of orchid evolution.
Although most orchids have a pollinium, Apostasia has powdery pollen, similar to Molineria capitulata, Oryza sativa (rice), and Arabidopsis thaliana. The P- and S-subclades of MIKC*-type genes are major regulators of powdery pollen8,9 and the loss of the P-subclade members of MIKC*-type genes has led to evolution of the pollinium. Formation of the pollinium, which is specialized for transfer as a single unit by pollinating vectors, was a key innovation in the evolutionary history of Orchidaceae, and may have played a role in promoting the tremendous radiation of orchids10.
A previous study suggests that type I Mb MADS-box genes regulate the development of endosperm7. In orchids, loss of this type of gene has led to seeds without endosperm. The evolution of seeds without endosperm has enabled orchids to transfer energy to the production of tens of thousands of seeds in a single fruit. Moreover, smaller seeds are more likely to disperse into new habitats and eventually lead to the formation of new species.
Orchids are one of very few flowering plant groups that have been able to successfully colonize epiphytic or lithophytic niches. We found that the AGL12 gene, which is involved in root cell differentiation11, regulates the root of Apostasia only when it is growing underground and absorbing nutrients. When orchids lack the AGL12 gene, lateral roots appear, which facilitate growth on trees or rocks. By exploiting habits that other plants cannot, this lifestyle enhances orchid growth and survival. There are large differences between these habitats, which can facilitate partition of gene exchange in orchid species (also known as habitat isolation) and thereby promote the formation of new species.
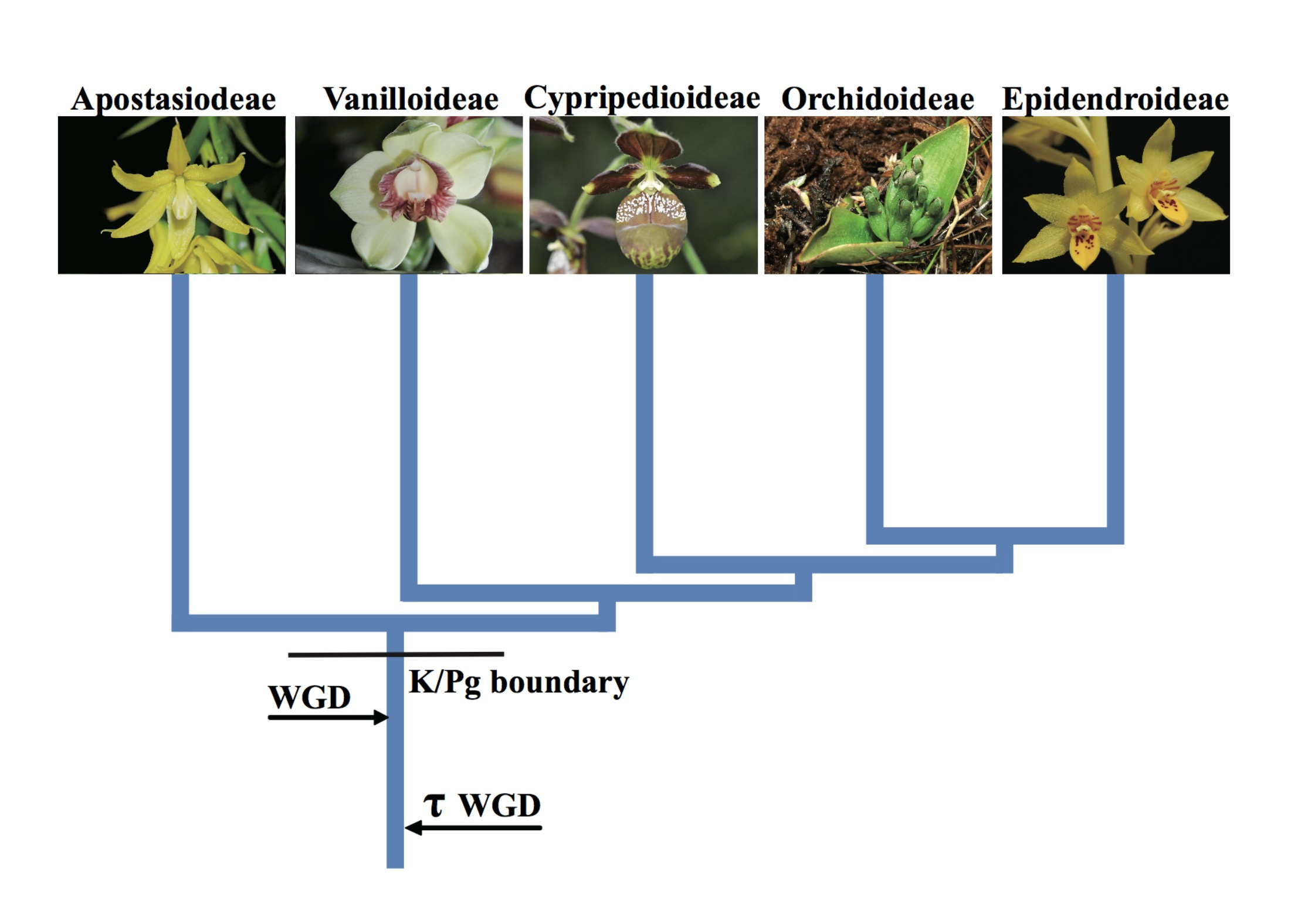
Figure 4 A WGD is shared by all orchids and occurred shortly before their divergence.
Perspectives
To the best of our knowledge, this study is the first to completely reconstruct the ancestral orchid gene toolkit and provides a route map of orchid evolution. Furthermore, we also reveal the molecular mechanisms of orchid flower development, which has permitted revision of the common knowledge of orchid evolution. In addition, our findings will provide an important theoretical basis and guidance for orchid protection and functional research.
References
1. Roberts, D. L. & Dixon, K. W. Orchids. Curr. Biol. 18, R325–R329 (2008).
2. Givnish, T. J. et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B 282, 1553 (2015).
3. Givnish, T. J. et al. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr. 43, 1905–1916 (2016).
4. Darwin, C. On the Various Contrivances by Which British and Foreign Orchids are Fertilised by Insects (Cambridge University Press, 2011).
5. Zhang, G. Q. et al. The Apostasia genome and the evolution of orchids. Nature, DOI 10.1038/nature23897. (2017).
6. Van de Peer, Y., Mizrachi, E., & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. online (2017).
7. Cai, J. et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72 (2015).
8. Liu, Y. et al. Functional conservation of MIKC* -Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288–1303 (2013).
9. Kwantes, M., Liebsch, D. & Verelst, W. How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302 (2012).
10. Johnson, S. D. & Edwards, T. J. The structure and function of orchid pollinaria. Plant Syst. Evol. 222, 243–269 (2000).
11. Tapia-López, R. et al. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182–1192 (2008).
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in