How microbial teamwork keeps plant healthy
Published in Ecology & Evolution, Microbiology, and Agricultural & Food Science

Phytopathogens are widespread over the world, constantly threatening plant health. They cause devastating plant diseases, leading to the loss of approximate 30% or even more of global crop yields each year and threatening food security worldwide1. Beyond yield loss, phytopathogens also undermine soil biodiversity and ecosystem stability, making soil health deterioration a global concern2. Yet, an interesting phenomenon is that even under the same field conditions, some plants remain remarkably healthy (Figure 1). What protects them? This simple question has fascinated us for years and guided our exploration into the invisible world beneath plant roots, the rhizosphere.

The rhizosphere, the narrow zone of soil surrounding plant root, influenced by root exudates, is a hotspot of microbial activity. Rhizosphere microbes not only contribute to nutrient cycling and plant growth, but also form a living barrier against invading pathogens3. Our global meta-analysis showed that higher microbial diversity in the rhizosphere rather than bulk soil constrain pathogen invasion4. However, the underlying mechanisms explaining trophic interactions between diverse microbial community and pathogen invasion remained unclear.
Our earlier study published in Nature Communications (2015, 6:8413) revealed that trophic networks representing substrate competition determine pathogen suppression: beneficial microbes compete with pathogens for carbon resources in the rhizosphere, effectively preventing pathogen invasion5. However, microbial interactions are far more complex than competition. Within microbial communities, there is also metabolite exchange or cross-feeding allowing microbes supporting each other’s growth (Figure 2). The importance of cross-feeding metabolites reused within microbial community on pathogen invasion is overlooked.
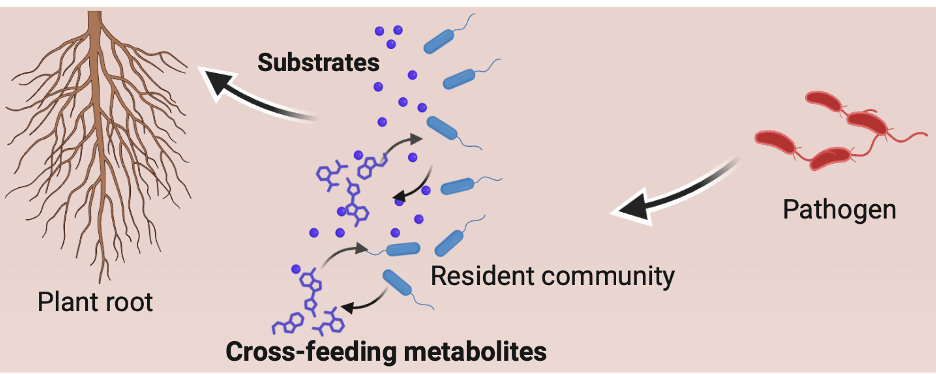
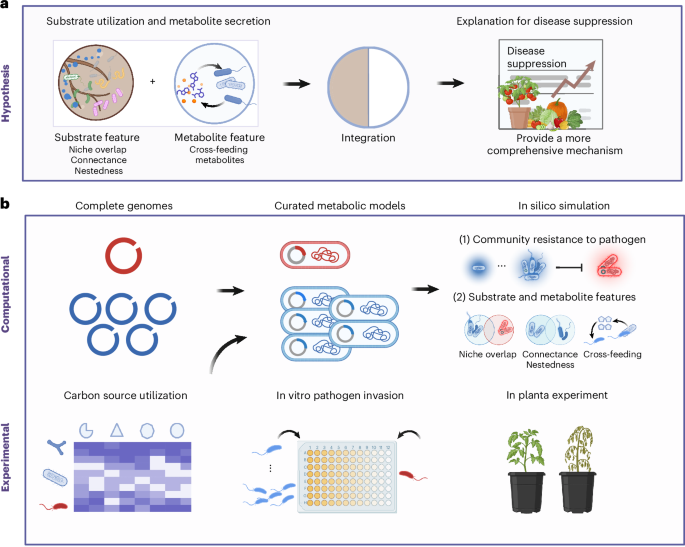
Predicting trophic interactions in multispecies communities has long been limited by experimental challenges. In vitro experiments designed to examine substrate and metabolite features are already labor intensive under controlled conditions and have become increasingly impractical in complex natural settings, where species composition and environmental conditions are highly variable. In this study, we combined genome-scale metabolic models with synthetic community experiments, both in vitro and in planta conditions. The models allowed us to simulate how microbes interact through nutrients and metabolites, and to predict community-level resistance under different nutritional environments in high efficiency and precision. This integrative approach bridges genomic data with metabolic ecology, enabling us to move from who is there to what they do and how they interact.
Our findings show that substrate utilization and cross-feeding synergistically determine microbiome resistance to pathogen invasion. Communities that combine strong external nutrient competition with active internal cooperation are the most robust against pathogen invasion. This new perspective provides a more comprehensice view on microbial interaction and highlights the importance of cooperative metabolism in maintaining soil and plant health.
Beyond the conceptual advances, our approach also demonstrates the power of genome-scale metabolic modeling for microbiome engineering. By predicting microbial interactions based on genomic and metabolic data, we are able to design disease-suppressive microbial communties tailored to specific crops regarding to specific rhizosphere resources. Such predictive frameworks could allow us to regulate rhizosphere microbiome more precise with certain carbon resources (e.g. rhizosphere prebiotic) — from reactive disease control to proactive microbial design.
Together, this work advances our understanding of how microbial trophic interactions determine plant health. It opens the door to microbiome-based strategies that enhance soil health, protect crops, and ultimately promote sustainable agriculture.
Reference
- Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3, 430–439 (2019).
- Singh, B. K. et al. Plant pathogens, microbiomes, and soil health. Trends in Microbiology https://doi.org/10.1016/j.tim.2025.03.013 (2025).
- Berendsen, R. L., Pieterse, C. M. J. & Bakker, P. A. H. M. The rhizosphere microbiome and plant health. Trends in Plant Science 17, 478–486 (2012).
- Yang, X. et al. High microbiome diversity constricts the prevalence of human and animal pathogens in the plant rhizosphere worldwide. One Earth 7, 1301–1312 (2024).
- Wei, Z. et al. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nature Communications 6, 8413 (2015).
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in