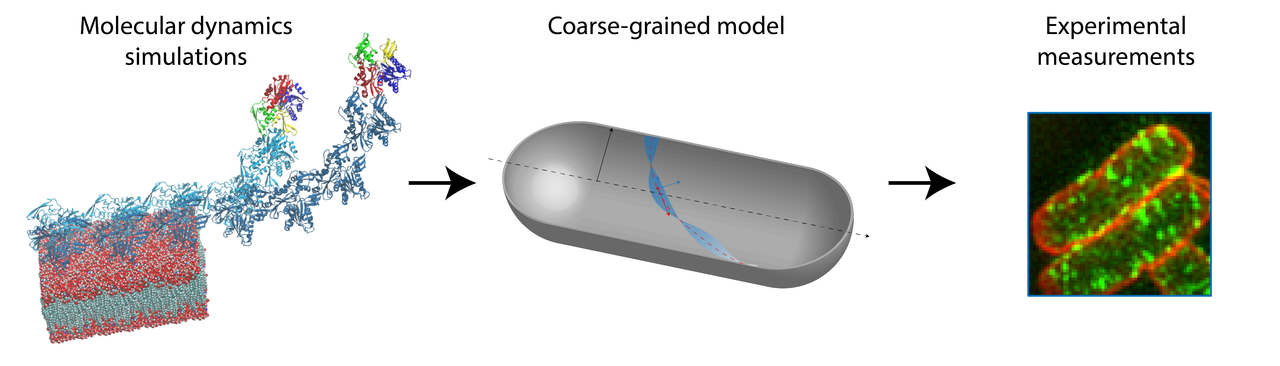
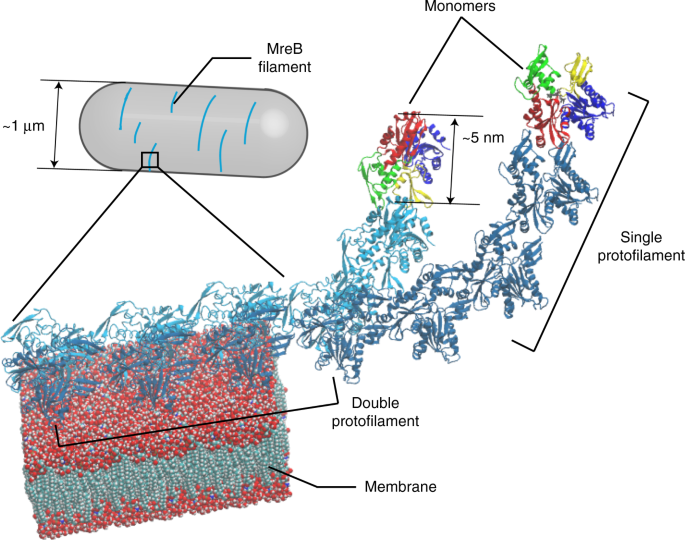
Bacterial cells maintain their shape and size surprisingly well during growth and proliferation. In many rod-shaped bacteria, cell shape is regulated by a homolog of the eukaryotic cytoskeletal protein actin, MreB (1). How does this small protein, with a size of ~5 nm, sense and maintain the geometry of a cell, which is several hundred-fold times larger? In our recent work, we combined molecular dynamics (MD) simulations, coarse-grained modeling, and experimental measurements to show that the size and conformation of MreB filaments within cells is predicted by molecular-level twist, highlighting a potential mechanism of MreB geometry sensing.
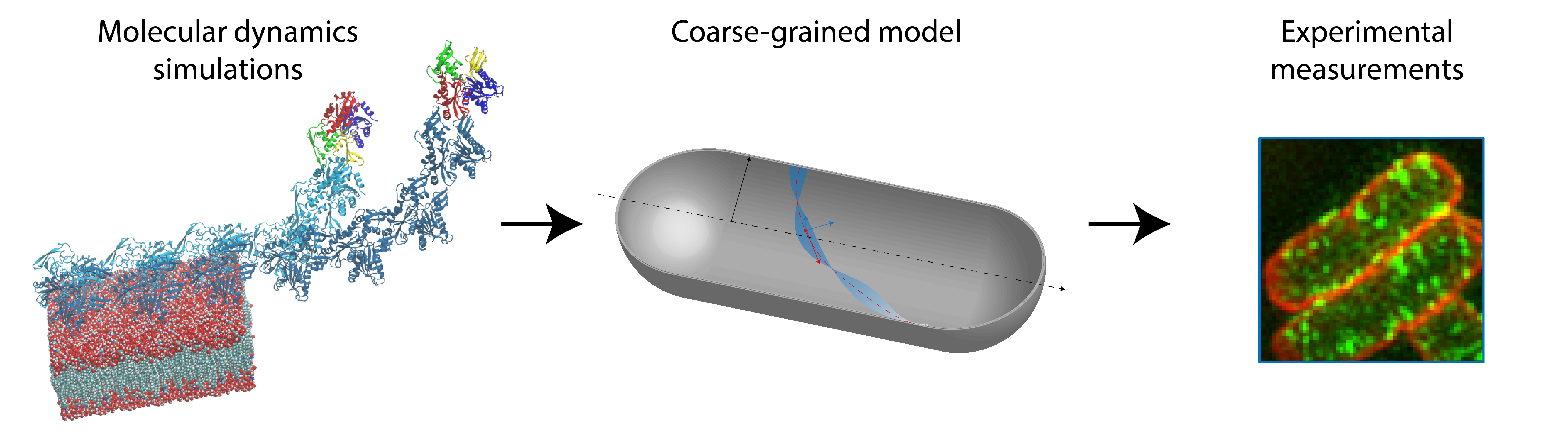
Figure 1. The combination of modeling and experiments allow accurate predictions of MreB filament conformations. We extracted kinetic parameters from MD simulations (left) to build a coarse-grained model (middle) of MreB filaments bound to a cylindrical membrane surface. The coarse-grained model predicted filament length and orientation based on their twist, which was later verified experimentally through single-cell super-resolution microscopy (right).
We started this work by performing all-atom MD simulations of MreB. Although MreB double protofilaments adopted a straight conformation in the crystal structure (2), in the simulations, when they were released from their crystal contacts we identified for the first time that they exhibited left-handed twist when surrounded by water molecules. Interestingly, when the protofilaments were placed close to a membrane patch, the twist was largely suppressed, allowing the MreB filaments to bind to the membrane surface. Other factors, such as point mutations in MreB and binding of a regulatory protein, also modulated twist.
A coarse-grained biophysical model had been developed by a group of theoretical physicists, Dr. Ajay Gopinathan, Dr. Gregory Grason, and Dr. David Quint (3), to address the properties of an infinitely long filament with intrinsic twist as it binds to a membrane: briefly, untwisting of such filaments builds up torsional energy, which is offset by the energy reduction due to membrane binding. This picture is highly consistent with our MD observations that MreB untwists during membrane binding. In collaboration with these physicists, we built a new coarse-grained model to accurately compute the interplay between these two energetic changes for a finite-long MreB filament on a cylindrical membrane surface, and found that the twist of MreB filaments was correlated with filament limit length and its relative orientation to the cylinder. Thus, we were able to predict that MreB mutants with different intrinsic twist would have different lengths and orientations in bacterial cells; we then experimentally measured the MreB filament length and orientation in the appropriate mutants with twist changes in our MD simulations and showed that the conformations of MreB filaments in vivo matched our model predictions.
Taken together, our work connects length scales from atoms to single cells, and reveals critical properties of MreB filaments. We postulate that the intrinsic twist of MreB filaments not only dictates their conformations, but also modulates the geometric preference of MreB. Such geometric preference potentially allows MreB to localize in certain areas of the cell body and promote cell wall synthesis to maintain rod-like cell shape (4). Our approach therefore provides the opportunity to engineer cell shape by directly tuning MreB properties in silico. Our coarse-grained model can also be modified to include the dynamics of MreB filaments, which will further depict polymerization as well as membrane binding dynamics of MreB, and connect such changes to cellular physiology.
Moving forward, the chiral twisting of MreB also provide a source of cellular chirality at the molecular level. Chirality in bacteria has been observed during growth of single cells (5) as well as in microbial biofilms (6). Thus, designing MreB mutants with different twists has the potential to understand and engineer chirality in bacterial cells.
References
- F. van den Ent, L. A. Amos, J. Löwe, Prokaryotic origin of the actin cytoskeleton. Nature 413, 39-44 (2001).
- F. Van den Ent, T. Izoré, T. A. Bharat, C. M. Johnson, J. Löwe, Bacterial actin MreB forms antiparallel double filaments. Elife 3, e02634 (2014).
- D. A. Quint, A. Gopinathan, G. M. Grason, Shape selection of surface-bound helical filaments: biopolymers on curved membranes. Biophysical journal 111, 1575-1585 (2016).
- T. S. Ursell et al., Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proceedings of the National Academy of Sciences 111, E1025-E1034 (2014).
- C. Tropini et al., Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell reports 9, 1520-1527 (2014).
- L. Jauffred, R. M. Vejborg, K. S. Korolev, S. Brown, L. B. Oddershede, Chirality in microbial biofilms is mediated by close interactions between the cell surface and the substratum. The ISME journal 11, 1688 (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in