IF1 governs the active/inactive state transitions of the sluggish ATP synthase.
Published in Cell & Molecular Biology

The mitochondrial ATP synthase is known as “the enzyme of life” because it provides most of cellular energy by oxidative phosphorylation (OXPHOS), but its essential role in cell physiology goes beyond that. The ATP synthase also plays a critical role in mitochondrial ultrastructure and function by its associating into dimers and oligomers that bend the inner mitochondrial membrane1. The regulation of ATP synthase activity has been extensively studied. However, the existence of an inactive pool of the enzyme in conditions of ATP production was largely ignored in the literature despite Moyle and Mitchell, many years ago, showed that mitochondrial preparations contain a fraction of inactive or “sluggish” enzyme that could transition to an active state2. Recent advances in super-resolution microscopy revealed functional heterogeneity in ATP synthase molecules, suggesting the existence of two subpopulations of the enzyme with different mobility and catalytic features3. In this work4, we have demonstrated that a pool of inactive ATP synthase co-exists with the active enzyme under normal OXPHOS conditions and identified the protein responsible for the transition between both states, the ATPase Inhibitory Factor 1 (IF1).
IF1 is a small protein that reversibly binds to the ATP synthase and blocks rotatory catalysis. It has been generally accepted that IF1 exclusively inhibits the reverse ATP hydrolytic activity of the enzyme to prevent the hydrolysis of ATP by de-energized mitochondria5, thus limiting its functional and physiological relevance in mitochondrial activities. In contrasts, previous studies from our lab have shown that overexpression of IF1 in cells in culture or in specific tissues in mice reduce the mitochondrial production of ATP, supporting that IF1 also inhibits the forward activity of the enzyme in energized mitochondria6-8. Notably, IF1 expression is up-regulated in human carcinomas and mesenchymal stem cells, contributing to metabolic reprogramming to the enhanced glycolytic phenotype that supports cellular proliferation by inhibition of OXPHOS6,9-11. More recently, we have shown in tissue-specific conditional mouse models that the genetic ablation of IF1 increases the production and turnover of ATP in mitochondria, confirming that IF1 acts as a “brake” to the enzyme under basal OXPHOS conditions8,12.
In this contribution4, we have confirmed both in cells in culture (Fig. 1a) and in the mouse tissues that express IF1, that mitochondria contain a pool of inhibited ATP synthase by its interaction with the endogenous IF1, as assessed by proximity ligation and co-immunoprecipitation assays (Fig. 1b, c), thereby reducing the fraction of active enzyme that can produce ATP (Fig. 1a). We uphold that the inhibited ATP synthase is the in vivo representative of the “sluggish” enzyme2 and support that it represents a reservoir of ATP synthase for the fine tuning between energy provision and expenditure, becoming activated upon a rapid increase in energy demand7. We found that the genetic ablation of IF1 impairs the formation of super-assemblies of the ATP synthase (Fig. 1d, e) and alters cristae organization (Fig. 1f), highlighting the critical relevance of IF1/ATP synthase for mitochondrial ultrastructure and function.
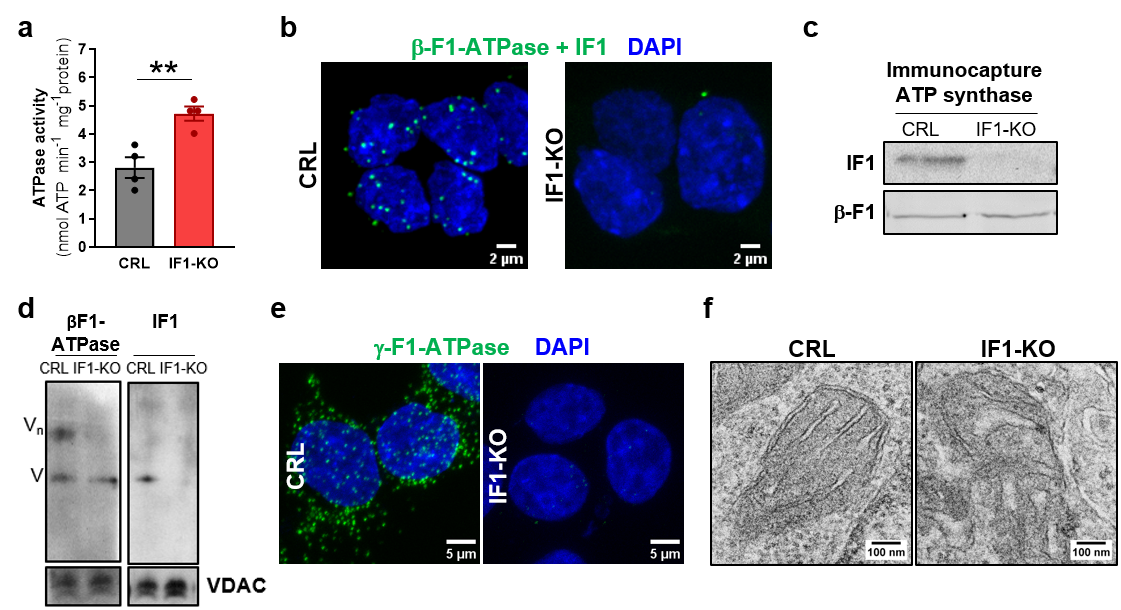
Figure 1. IF1 controls a pool of inactive ATP synthase in mitochondria. (a), Histograms showing the hydrolytic activity of ATP synthase in CRL and IF1-KO HCT116 cells. ** p≤0.01 when compared to CRL by t-Student’s test. (b), Representative images of Proximity Ligation Assay showing the interaction between β-F1-ATPase and IF1 (green dots) in CRL and IF1-KO HCT116 cells. (c), Immunocapture of ATP synthase from mitochondrial extracts of CRL and IF1-KO HCT116 cells blotted against IF1 and β-F1-ATPase. (d), Representative BN-immunoblots of complex V (β-F1-ATPase and IF1) in isolated mitochondria from CRL and IF1-KO HCT116 cells. VDAC is shown as loading control. (e), Representative images of Proximity Ligation Assay using g-F1-ATPase as target (green dots) in CRL and IF1-KO cells. (f), Representative electron micrographs of mitochondria from CRL and IF1-KO HCT116 cells.
By consuming the proton electrochemical gradient generated by the electron transport chain, the ATP synthase modulates mitochondrial membrane potential (Dym) in OXPHOS. Hence, IF1-mediated inhibition of the enzyme promotes an increase in Dym6,8. However, does this occur evenly throughout each mitochondrion or rather in specific cristae microdomains? To answer this question, we developed a fusion construct of IF1 with green fluorescent protein (GFP) that allowed us to observe by super-resolution microscopy the intramitochondrial distribution of IF1 in live cells. Importantly, we found cristae microdomains with high IF1 localization that also have high Dym, highlighting that local regulation of the ATP synthase by IF1 contributes to the heterogeneity of Dym in different cristae from the same mitochondrion. It should be noted that a construct with a mutant version of IF1-GFP, unable to bind to the ATP synthase, or GFP targeted to mitochondria, showed no co-distribution between GFP and high Dym, supporting that the interaction of IF1 with the enzyme determines the local active/inactive state transitions of the sluggish ATP synthase.
At an ultrastructural level, electron microscopy imaging of immunolabeled tissue slices revealed that IF1 and ATP synthase also co-distribute in mitochondria from mouse tissues, with a relevant fraction of the enzyme in close proximity to the inhibitor. Furthermore, we found a preferential localization of the ATP synthase at cristae tips, supporting that IF1 interaction is likely to stabilize the rows of ATP synthase in this location to maintain cristae structure.
In summary, we have found two pools of ATP synthase in mitochondria in vivo, active and inactive or “sluggish”, being the transition between both states controlled by the interaction with IF1, which is also critical for the formation of super-assemblies of ATP synthase at cristae tips. Moreover, the interaction is regulated at the local level, which poses exciting new questions: how local factors control IF1 interaction with the ATP synthase? Do different cristae microdomains fulfill distinct roles, for example ATP production and/or signaling by changes in Dym and the production of reactive oxygen species? Since IF1 is not expressed in all tissues, the development of tissue-specific mouse models of loss and gain of function of IF1 will be key to portray the multifaceted roles of IF1/ATP synthase in tissue homeostasis and pathology. Overall, helping to understand mitochondrial structure and function.
Full text available at: https://www.nature.com/articles/s42003-023-05214-1
References:
1 Kuhlbrandt, W. Structure and Mechanisms of F-Type ATP Synthases. Annu. Rev. Biochem. 88, 515-549, doi:10.1146/annurev-biochem-013118-110903 (2019).
2 Moyle, J. & Mitchell, P. Active/inactive state transitions of mitochondrial ATPase molecules influenced by Mg2+, anions and aurovertin. FEBS Lett. 56, 55-61 (1975).
3 Weissert, V. et al. Inhibition of the mitochondrial ATPase function by IF1 changes the spatiotemporal organization of ATP synthase. Biochim Biophys Acta Bioenerg 1862, 148322, doi:10.1016/j.bbabio.2020.148322 (2021).
4 Romero-Carraminana, I., Esparza-Molto, P. B., Dominguez-Zorita, S., Nuevo-Tapioles, C. & Cuezva, J. M. IF1 promotes oligomeric assemblies of sluggish ATP synthase and outlines the heterogeneity of the mitochondrial membrane potential. Commun Biol 6, 836, doi:10.1038/s42003-023-05214-1 (2023).
5 Walker, J. E. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1-16, doi:10.1042/BST20110773 (2013).
6 Sanchez-Cenizo, L. et al. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 285, 25308-25313, doi:10.1074/jbc.M110.146480 (2010).
7 Garcia-Bermudez, J. et al. PKA Phosphorylates the ATPase Inhibitory Factor 1 and Inactivates Its Capacity to Bind and Inhibit the Mitochondrial H(+)-ATP Synthase. Cell Rep 12, 2143-2155, doi:10.1016/j.celrep.2015.08.052 (2015).
8 Esparza-Molto, P. B. et al. Generation of mitochondrial reactive oxygen species is controlled by ATPase inhibitory factor 1 and regulates cognition. PLoS Biol. 19, e3001252, doi:10.1371/journal.pbio.3001252 (2021).
9 Formentini, L., Sánchez-Aragó, M., Sánchez-Cenizo, L. & Cuezva, J. M. The mitochondrial ATPase Inhibitory Factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol. Cell 45, 731-742, doi:10.1016/j.molcel.2012.01.008 (2012).
10 Sanchez-Arago, M., Garcia-Bermudez, J., Martinez-Reyes, I., Santacatterina, F. & Cuezva, J. M. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep 14, 638-644, doi:10.1038/embor.2013.72 (2013).
11 Dominguez-Zorita, S. & Cuezva, J. M. The Mitochondrial ATP Synthase/IF1 Axis in Cancer Progression: Targets for Therapeutic Intervention. Cancers (Basel) 15, doi:10.3390/cancers15153775 (2023).
12 Dominguez-Zorita, S. et al. IF1 ablation prevents ATP synthase oligomerization, enhances mitochondrial ATP turnover and promotes an adenosine-mediated pro-inflammatory phenotype. Cell Death Dis. 14, 413, doi:10.1038/s41419-023-05957-z (2023).
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in