Getting RIDD of insect pests: Targeting doublesex to make dominant sterile CRISPR homing gene drive
Published in Protocols & Methods, Sustainability, and Agricultural & Food Science
Pest control plays a critical role in safeguarding public health, ensuring food security, and protecting ecosystems. Pests cause significant damage to crops, lead to the spread of diseases, and disrupt natural habitats. Effective pest management strategies are essential to prevent economic losses in agriculture and to reduce the risks associated with pests.
One promising approach for pest control is the use of genetic engineering technology, particularly gene drive systems. Gene drive refers to a mechanism that increases the likelihood of a specific gene being passed on to the next generation, thereby spreading desirable traits rapidly within a population. This powerful tool has the potential to enhance pest control efforts by altering pest populations at a genetic level, reducing their ability to reproduce or survive, and ultimately leading to a decrease in their numbers.
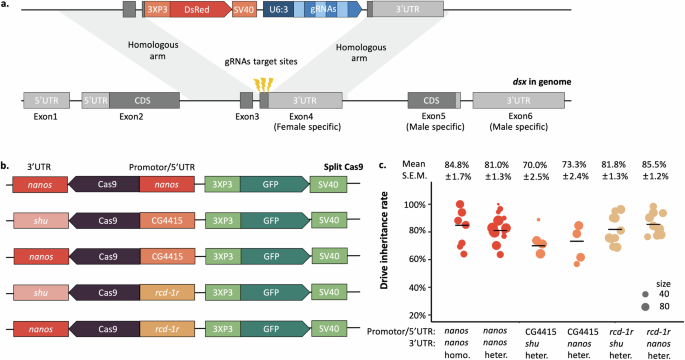
The most commonly studied gene drive system is the CRISPR homing drive, which converts wild-type alleles into drive alleles in the germline of drive heterozygotes. Cas9 and gRNA cleave the wild-type allele, which then undergoes homology-directed repair. We call this process ‘drive conversion’ or 'homing', and it increases the proportion of drive alleles among the offspring. However, DNA repair can also take place by end-joining, introducing mutations at the target site and thus creating resistance alleles that Cas9 cannot recognize. Resistance alleles will accumulate in the population, block the spread of drive, and prevent population elimination, which is a significant challenge for current CRISPR homing drives.
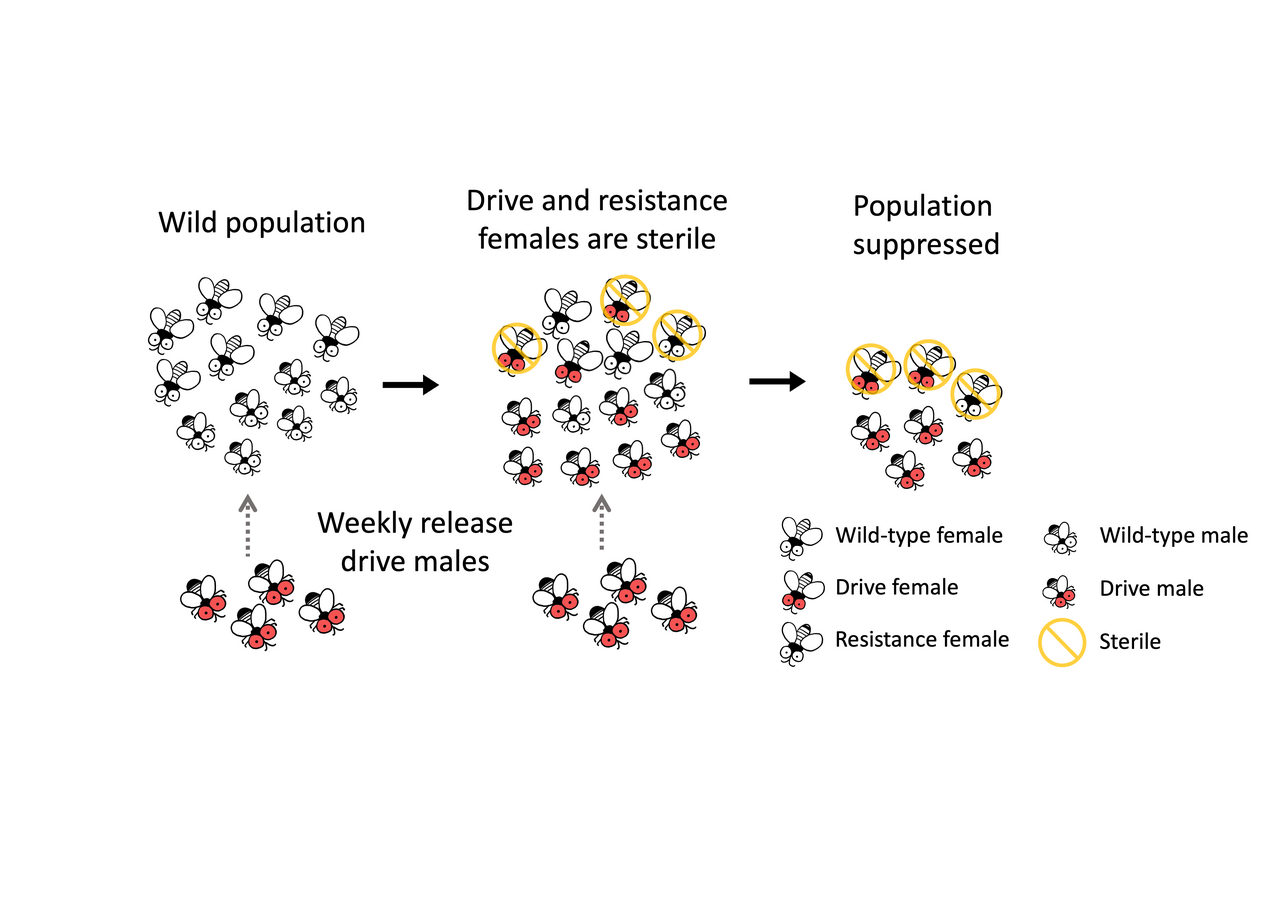
In this study, we wanted to develop a CRISPR-based homing suppression gene drive targeting the female exon of doublesex in Drosophila melanogaster. doublesex is an important but haplosufficient gene for sex determination of Diptera, with sex-specific splicing. We choose a female specific site as the target, similar to Kyrou et al. 2018 and Yadav et al. 2023, but with multiple gRNAs to reduce the chance of functional resistance. We expected all homozygous drive females to be sterile, which is desirable for population suppression gene drive. If the drive is efficient enough and avoids functional resistance, the population should be eliminated after the drive allele spreads sufficiently widely.
However, we found that all of our heterozygous drive females were sterile and masculinized. This was unexpected. We were upset at first because the dominant-sterile drive will be rapidly removed from the population in drive heterozygous females, preventing the drive from increasing in frequency in the population. However, the drive still works in males, so we estimated that we could still suppression the population if we repeated release drive males, similar to the well-studied Sterile Insect Technique (SIT).
Interestingly, we also found that resistance allele were dominant female sterile, which means that our system can quickly eliminate resistance alleles from the population. doublesex mutations leading to dominant female sterility were originally reported in the 1990s, but our system was able to consistently generate a variety of dominant sterile alleles using its three gRNAs. Dominant resistance improved the suppression efficiency. We then realized this drive system will not be something of little interest. It could actually represent a new strategy that allows more powerful but self-limiting suppression. In this system, heterozygous drive males spread drive and resistance alleles, keep generating dominant sterile females. Male progeny can continue to spread the drive, or at least resistance alleles. As a result, the population will gradually lose its fertility. To prove that this strategy should be more efficient than previous genetic pest control approaches, we simulated SIT, Female-specific Release of Insects carrying a Dominant Lethal gene (fsRIDL), and our new drive system, which required far fewer transgenic males released for suppression.
To thoroughly demonstrate the feasibility of our drive system, we designed and conducted an overlapping-generation cage study. By releasing heterozygous males weekly, we successfully suppressed the populations in two cages, each starting with over 2,000 flies, within approximately 30 weeks. Our experimental design had five food bottles in each cage, and we replaced the oldest food with a fresh bottle every three days. We tracked the population size, and we utilized an aspirator to sample and screen flies, allowing us to monitor both drive frequency and sex ratio. The cage populations were structured by age, closely resembling natural wild populations. This type of cage study can be used for investigating other pest control strategies in the future, particularly those that involve repeated releases.
Lab website: https://jchamper.github.io/
Reference:
Kyrou, K., Hammond, A.M., Galizi, R., Kranjc, N., Burt, A., Beaghton, A.K., Nolan, T., and Crisanti, A. (2018). A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol 36, 1062–1066. https://doi.org/10.1038/nbt.4245.
Yadav, A.K., Butler, C., Yamamoto, A., Patil, A.A., Lloyd, A.L., and Scott, M.J. (2023). CRISPR/Cas9-based split homing gene drive targeting doublesex for population suppression of the global fruit pest Drosophila suzukii. Proceedings of the National Academy of Sciences 120, e2301525120. https://doi.org/10.1073/pnas.2301525120.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in