In Pursuit of Single-Cell Insights of Organoid Signalling
Published in Protocols & Methods

During the last decade, developments in organoid technology have revolutionised the ways scientists can study complex biological systems. Driven by stem cell proliferation and differentiation (and most likely local cell-cell communication), organoids demonstrate remarkable self-organisation and resemblance to their tissue of origin.
While complexity is what make organoids so powerful, it also makes organoids incredibly challenging to study. Although holistic approaches such as viability measurement and whole-mount imaging have provided great insights into organoids as a heterogeneous population, such methods will inevitably err on the side of ignoring finer biological details where a high signal-to-noise ratio is key. In fact, as organoids are being used as building blocks for increasingly sophisticated culture systems, it has become even more critical that we can obtain cell type-specific, or better still, single-cell data from organoids. In our recent paper in Nature Methods, we expanded the current toolbox for organoid phenotyping with a newly developed method to study proteins and post-translational modifications (PTMs) in single organoid cells.
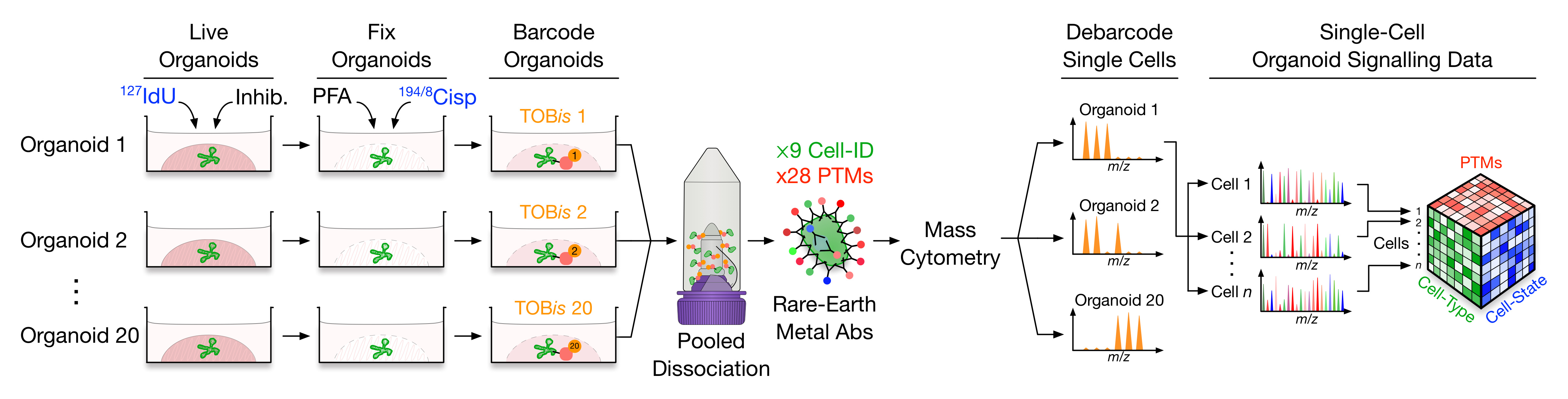
Figure 1, Experimental workflow. Live organoids are pulsed with IdU to label S-phase cells, treated with protease / phosphatase inhibitors and fixed with PFA to preserve post translational modification (PTM) signals, and stained with Cisplatin to label dead cells. Fixed organoids are barcoded using Thiol-reactive Organoid Barcoding in situ (TOBis), pooled for dissociation, stained with rare-earth metal-conjugated antibodies, and analysed by single-cell mass cytometry (MC). The resulting dataset contains integrated cell-type, cell-state, and PTM signalling information from multiple organoid culture conditions.
In a nutshell, our method took inspiration from the existing mass cytometry (MC) protocols designed for immunophenotyping and applied it to taking snapshots of organoid signalling at single-cell resolution (Figure 1). Looking back, the most important take-home message of the paper is perhaps the revelation of the intimate links between cell-type, cell-state, and signalling within organoids. Alternatively, one may argue that the direct comparison of oncogenic mutation- and stroma-driven organoid PTM profiles is something only made possible by the presented work. These are undoubtedly well-grounded discussions — the ones that are still ongoing in the lab — but it might be worthwhile to reflect on some of the thoughts that are not explicitly spoken of in the paper, yet have also been pondered on along the way.
During the development of the project, one of the design principles decided fairly early on was to put the fixation step upfront in the protocol. The rationale is straightforward: PTM signalling is notoriously sensitive to external cues and can undergo dramatic changes within seconds. The very act of dissociating organoids into single cells could easily destroy the PTM signalling profile. Fixing first could protect the PTMs. In retrospect, however, an early fixation is beneficial in many more ways beyond the preservation of biological signals. It is only with fixation that organoids can possibly withstand the subsequent dissociation and come out single cells in suspension (rather than a soup of cell debris), and an early snap-freeze minimises the risk of technical noises sneaking in during the time lag of sample handling — something of considerable impact particularly for large-scale experiments.
Talking about technical variability and experimental scalability, these are exactly the motivations that underpinned the development of our new multiplexed organoid barcoding strategy. Mass-tagged barcoding is well established in the MC community — we’ve got the luxury to allocate a number of mass channels for sample labelling and deconvolution. Unfortunately, the current barcoding methodology is designed to work with single cells in suspension (e.g. immune cells) and is not compatible with organoids embedded within Matrigel. We can still use the old technology to barcode organoids, but only after they have been dissociated into single cells and resuspended in a protein-free solution — one condition at a time. This is a huge pain in practice, but more importantly, it is not scalable to large experimental designs and clearly opens the door to technical variations. Not willing to give up on either scalability or reproducibility, we decided to work from the bottom up and design a barcoding strategy specifically for organoids. Thiol-reactive Organoid Barcoding in situ (TOBis) was developed as a result, paving the path for high-throughput organoid MC analysis, and in the meanwhile makes our life much easier!
On a final note, one mindset we’ve been constantly reminding ourselves not to fall into throughout the lifetime of the project is the trap of developing a method merely for the sake of method development. At the end of the day, we need a well-defined, legitimate biological question at its core, and a meticulously optimised experimental system as the touchstone and playground. In the case of this study, what we really wanted to know was how cells communicate in a heterogeneous multicellular environment, especially in cancer. We set the scene using organoid models for the colorectal cancer (CRC) tumour microenvironment, went into the efforts of fine-tuning cell ratios, seeding density, medium composition as well as culture duration for the experiment, and applied the newly developed MC workflow as the final step. Only then were we comfortable to say we’ve learned something new about cell signalling in CRC – back to where everything began. There is still a long way to go, but now we have the tools to finally answer these questions.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in