Incorporating the image formation process into deep learning improves network performance
Published in Bioengineering & Biotechnology

Fluorescence microscopy plays a key role in modern biological research. However, due to blurring and noise intrinsic to the imaging process, the raw fluorescence image data recorded by the microscope is always degraded. To achieve high resolution and rapid imaging, many labs have put significant effort into improving microscopy or the associated image processing algorithms. These efforts are often bolstered by collaboration. In my own case, I have had a fruitful collaboration with researchers in Dr. Hari Shroff’s lab at the National Institute of Biomedical Imaging and Bioengineering (NIBIB, now at Janelia Research Campus), Dr. Patrick La Riviere’s lab at the University of Chicago, and my advisor Dr. Huafeng Liu at Zhejiang University. In 2020, we introduced the concept of the ‘unmatched back projector’ into Richardson-Lucy deconvolution1, 2 (RLD) and also developed a deep learning method (DenseDeconNet, DDN) to accelerate the processing of microscopy datasets by orders of magnitude3 (see our previous Blog post: Rapid image deconvolution and multiview fusion for optical microscopy | Nature Portfolio Bioengineering Community).
In 2021, Drs. Jiji Chen and Hari Shroff and their collaborators used three-dimensional residual channel attention networks (RCAN) to denoise and enhance their fluorescence microscopy data4. Around the same time, Drs. Yicong Wu, Hari Shroff and collaborators synergized concepts from multiview imaging, structured illumination, and deep learning (RCAN and content-aware image restoration (CARE) networks5) to improve the performance of confocal microscopy6. Although these efforts highlighted the power of DDN, RCAN and CARE for image restoration, they also emphasized that the performance of these networks depend crucially on the quality of the training data, and that the networks had trouble generalizing to new types of data not seen during training. Therefore, we wondered if it might be possible to improve the generalization capability of deep learning, perhaps by efficiently incorporating the physical model of image formation into the learning-based method.
When Dr. Yicong Wu discussed these challenges with Dr. Huafeng Liu (they have known each other for 20 years since they worked together at Hong Kong University of Science and technology), Dr. Liu inspired me to seek a solution through algorithm unrolling7. My colleagues in Dr. Liu’s lab have investigated the algorithm unrolling methods ADMM-net8 and ISTA-net9 for positron emission tomography (PET) image reconstruction, which can learn key parameters used in traditional iterative algorithms. This made us wonder: could a similar approach learn the unmatched forward/back projectors we had previously introduced for RLD? An obvious similarity between RLD and a fully convolutional deep learning network is that the convolution operation plays an important role in both methods. Through proper design, the convolution with forward/back projectors in RLD can be replaced with convolutional layers used in a neural network. After a series of trial and error, I successfully created the structure used in Richardson-Lucy Network (RLN), incorporating the physical model of RLD into a fully convolutional network and enabling the network to use both a large field of view and fine image details (Figure 1).
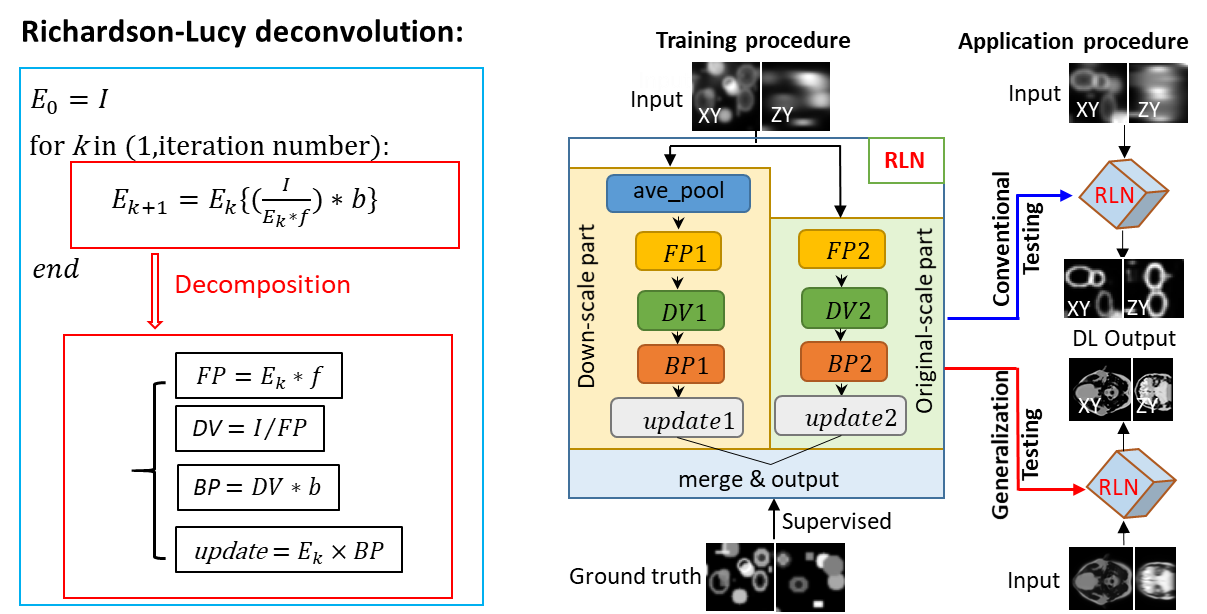
Figure 1. The decomposition of Richardson-Lucy deconvolution and the training and application of Richardson-Lucy Network. Schematic design of RLN consisting of three parts: down-scale part, original-scale part, and merge & output part. FP1, DV1, BP1, FP2, DV2, BP2 in down-scale part and original-scale part follow the RL deconvolution iterative formula. During training, low-resolution volumes (i.e., input) are fed into RLN, and the corresponding high-resolution ground truth are used to supervise the learning of network parameters. When applying the model, we perform conventional testing (training datasets include the same types of structures as in the test datasets) and generalization testing (i.e., test datasets are from different types of structures than the training datasets).
Next, I conducted tests of RLN on simulated data, demonstrating that some aspects of RLN can be easily interpreted and that it requires a relatively small training dataset to achieve excellent deconvolution capability equivalent to RLD. Drs. Yicong Wu and Hari Shroff and other collaborators (e.g., Daniel Colón-Ramos at Yale University) were excited with the preliminary results, and they designed real biological experiments on a diverse set of microscopes to better highlight the advantages of RLN vs. other deep learning methods. Through our joint efforts, we systematically demonstrated the advantages of RLN. For example, by combining RLN with a ‘crop and stitch’ module, we found it provided 4-6-fold faster processing of large cleared-tissue light-sheet data, e.g., for a volume of ~250 GB size (6370 x 7226 x 2850 voxels), RLN required ~3.7 hours vs. ~ one day with a previous processing pipeline3. We anticipate that RLN can be further tweaked to provide even faster deconvolution.
Perhaps the most exciting advantage of RLN is its excellent generalization ability. By training on synthetic data (Figure 2a), RLN provides robust deconvolution on real biological datasets that share the same PSF as the synthetic data. We found that RLN provided better output than traditional RLD (Figure 2b) and state-of-the-art commercial deconvolution software. We believe the ability to train on synthetic data is especially useful, particularly because it is not always possible to generate high SNR, high quality training ground truth.
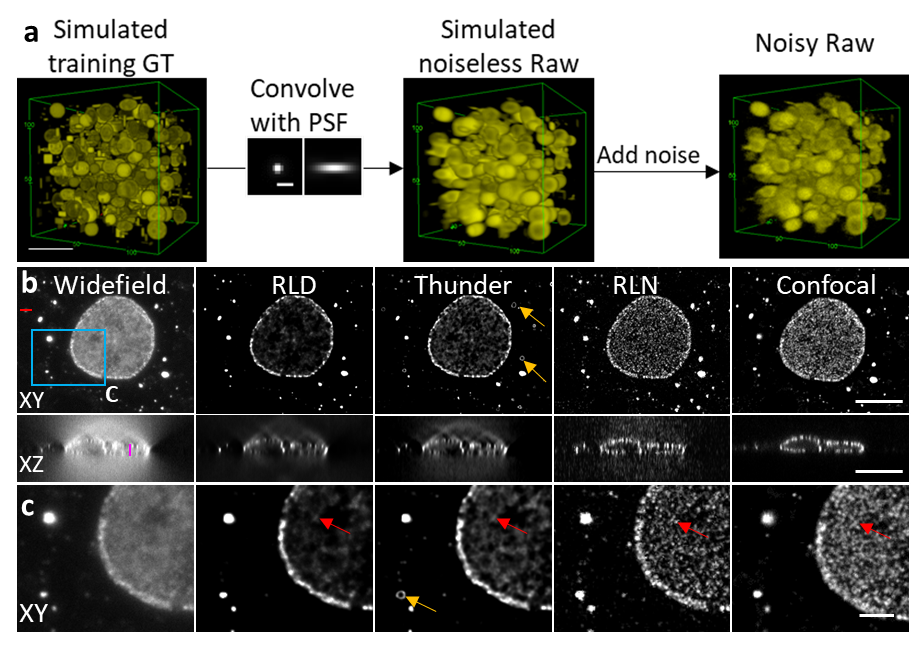
Figure 2. RLN trained with synthetic data can outperform RLD for datasets acquired by widefield microscopy. (a) Generation of synthetic data. (b) Lateral and axial planes of images of nuclear pore complexes in a fixed Cos-7 cell immunolabeled with primary mouse anti-Nup clone Mab414 and goat-anti-mouse IgG secondary antibody conjugated with Star635P, comparing widefield input, RLD with 100 iterations, Leica Thunder output, RLN prediction, and registered confocal data as a ground-truth reference. (c) Magnified views of the blue rectangle in (b). RLN was trained with synthetic mixed structures. Visual analysis (e.g., red arrows) demonstrate that RLN restoration outperforms Thunder (obvious artifacts indicated by orange arrows) and RLD in both lateral and axial views, showing detail that approaches the confocal reference. Scalebar: (a) 5 μm; (b) 10 μm; (c) 3 μm.
- Richardson, W.H. Bayesian-based iterative method of image restoration. JoSA 62, 55-59 (1972).
- Lucy, L.B. An iterative technique for the rectification of observed distributions. The astronomical journal 79, 745 (1974).
- Guo, M. et al. Rapid image deconvolution and multiview fusion for optical microscopy. Nature Biotechnology 38, 1337-1346 (2020).
- Chen, J. et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes. Nature methods 18, 678-687 (2021).
- Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nature methods 15, 1090-1097 (2018).
- Wu, Y. et al. Multiview confocal super-resolution microscopy. Nature 600, 279-284 (2021).
- Monga, V., Li, Y. & Eldar, Y.C. Algorithm Unrolling: Interpretable, Efficient Deep Learning for Signal and Image Processing. IEEE Signal Processing Magazine 38, 18-44 (2021).
- Yang, Y., Sun, J., Li, H. & Xu, Z. Deep ADMM-Net for compressive sensing MRI. In 30th international conference on neural information processing systems, 10-18 (NIPS, 2016)
- Zhang, J. & Ghanem, B. ISTA-Net: Interpretable optimization-inspired deep network for image compressive sensing. In 31st IEEE/CVF conference on computer vision and pattern recognition (CVPR), 1828-1837 (IEEE, 2018)
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in