Inferring HIV-1 transmission networks and sources of epidemic spread in Africa with deep-sequence phylogenetic analysis
Published in Microbiology

Two weeks ago, Dr. Fauci, head of NIAID, announced a campaign to eradicate HIV in the United States. The campaign rests on four pillars: diagnose all cases as early as possible, treat infections rapidly and effectively, prevent people at risk from being infected and rapidly detect and respond to fast-growing transmission networks, tracing the virus through the population to find individuals with unidentified infections and those at risk of being infected by the outbreak.
This is an ambitious project even in the United States where an estimated 160,000 are unaware that they are carrying the virus. Now imagine the same challenge in a country like Uganda, where an estimated 250,000 HIV-infected people are unaware of of their HIV status, or in South Africa, where this number is closer to 700,000 individuals. Tracing the virus from one infected person to the next in these settings is not just ambitious, it’s a Herculean task. This is because tracing requires individuals to name their sexual partners who in turn need to be found by health care workers.
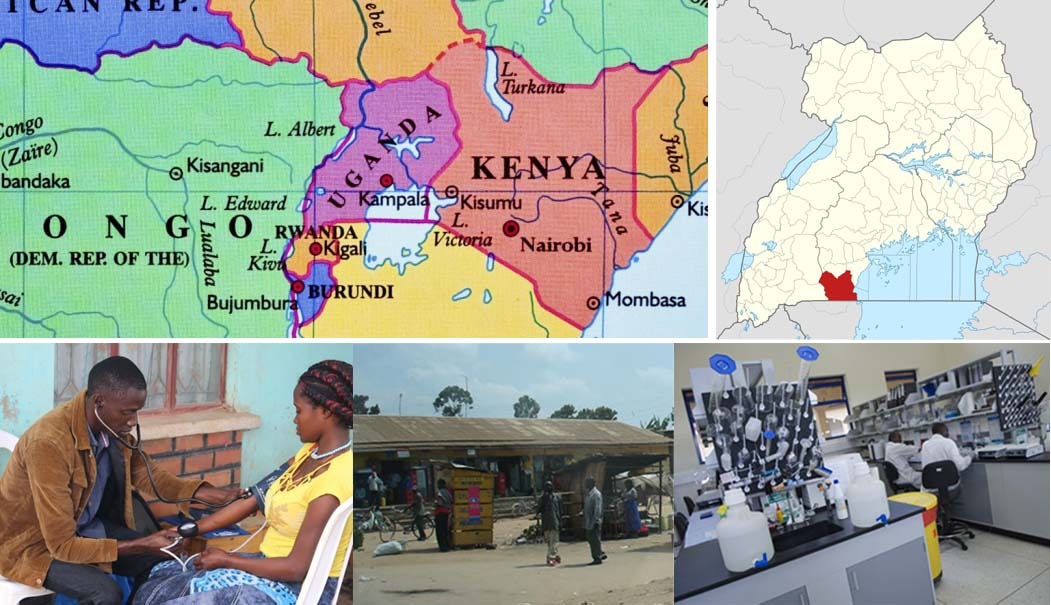
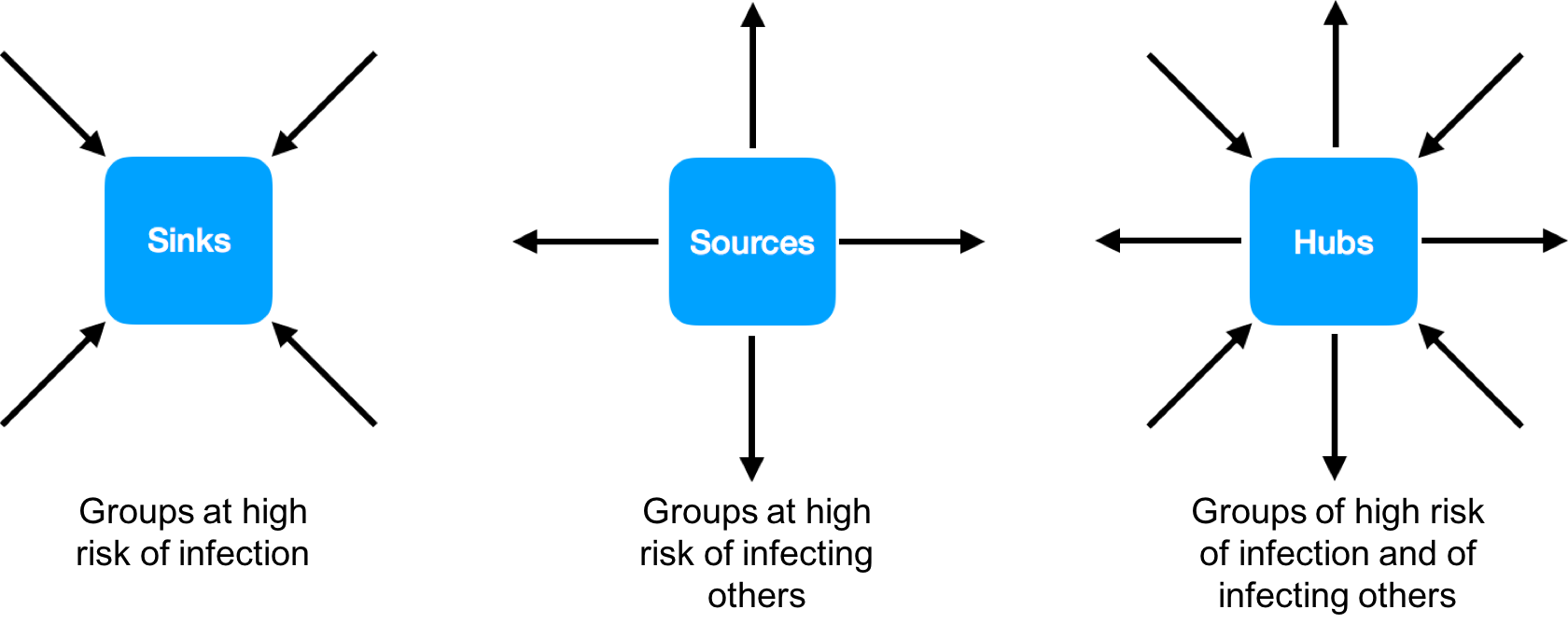
In contrast to tracing individuals, the epidemic can more broadly be described by a model of connected sources, sinks, and hubs. Sources are groups in a population that disproportionately pass on infections, sinks are groups that disproportionately receive infections, and hubs are both sources and sinks. Population groups can be defined by age and gender, or other characteristics and combinations thereof. Identifying these groups also allows prevention to be tailored. We reported last week on the use of viral deep sequence data to find groups of persons at high risk of transmitting HIV in the greater Rakai region of South central Uganda. Rakai was where the first cases of HIV were identified in Eastern Africa and was considered the epicentre of the HIV epidemic in the Lake Victoria region. Today, adult HIV prevalence is about ~13% in the Rakai community cohort; a cohort of ~20,000 individuals studied by the Rakai Health Sciences Program (RHSP) for nearly 25 years (Figure 1). However, in some of the cohort communities nearly half of the population is HIV-infected. In this cohort, over 80 percent of the HIV-infected individuals received antiretroviral treatment and over 70 percent are virally suppressed.
In collaboration with researchers at the RHSP in Uganda and the Bill and Melinda Gates funded PANGEA-HIV consortium, we used next generation sequencing methods to generate thousands of HIV sequences from HIV-infected persons in Rakai. Using these viral deep sequence data, we demonstrated that even against a high background of ongoing transmission, it is possible to construct HIV transmission networks. Crucially, new sequencing and analysis methods allow reconstruction of directed transmission networks. That is, the inferred networks are not just clusters of individuals with similar virus: they give insights into the direction of viral flow through the population. These clusters can be defined by age and gender, socioeconomic factors, riskiness of sexual behaviour, geographical location or any other relevant attributes. The method therefore allows the identification of characteristics that put certain groups within a population at high risk of acquiring and/or transmitting HIV (Figure 2).
As expected, high viral load in the transmitter increased transmission from both men to women and women to men. Men who reported more than one female partner in the last year were more likely to pass on the virus, however a similar relationship was not observed for transmission from women to men. In many communities, men were twice as likely to pass on the virus within the household than women. Future work within the PANGEA consortium will look at more epidemiological factors in different regions in sub-Saharan Africa.
Inferring transmission with direction has ethical and legal implications. Our approach cannot unequivocally prove that one individual infected another individual – the study provides several counterexamples – but the inferences are good enough to identify viral flow between different groups within the population. Furthermore, the work was conducted under the PANGEA accreditation system, where sharing of sensitive data is limited to researchers who abide by a strict code of conduct to maintain the anonymity of study participants.
Analyses of this kind will complement the many lessons already learned from epidemiologic studies in the fight to end the HIV/AIDS epidemic. Antiretroviral therapy has transformed HIV from a fatal disease to a chronic, manageable condition. Infection rates are on the decline, thanks to major breakthroughs in HIV prevention, including free and voluntary HIV testing services, early HIV treatment, initiation voluntary male medical circumcision, pre-exposure prophylaxis, condom provision, education and empowerment. The tools to control the pandemic are available. The challenge is comprehensive and strategic implementation in the US and international eradication programmes and beyond, for which identifying the groups at high risk of acquiring and transmitting the virus will be key.
Post written by the study authors, with special thanks to Lucie Abeler-Dörner who conceived the text on a late night flight.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in