Integrating genetics and proteomics to identify novel causal and druggable proteins for Parkinson’s disease
Published in Neuroscience

Parkinson’s disease (PD) is the 2nd most common neurodegenerative disease (after Alzheimer's disease), affecting dopaminergic neurons in the substantia nigra and leading to motor symptoms. LRRK2 (leucine-rich repeat kinase 2) is involved in lysosomal trafficking and degradation and has multiple common genetic variants associated with PD risk. Microglia and immune system function has been shown to be important in the underlying pathobiology of PD, with impaired autophagy affecting lysosomal degradation of proteins such as α-synuclein. With mutations linked to both familial (autosomal dominant) and sporadic PD, understanding the involvement of protein interactions and pathways will pave the way for LRRK2-targeted therapies.
In order to understand the downstream processes leading to disease due to LRRK2 genetic variants, we decided to determine if those same LRRK2 variants are also associated with cerebrospinal fluid (CSF) protein levels, and what biological pathways those proteins belong to. In fact, our previous studies in a small dataset found that the LRRK2 variants were associated with CSF GRN (granulin), another known protein involved in neurodegeneration and PD and we hypothesize other proteins may be also associated with LRRK2. To do this, we used the largest aptamer-based CSF proteomics study to date (7,006 proteins in 3,107 individuals) and performed comprehensive proteogenomic analyses to identify proteins associated with LRRK2. We then used additional mediation and pathway analyses to better understand downstream effects of the identified proteins (Figure 1).
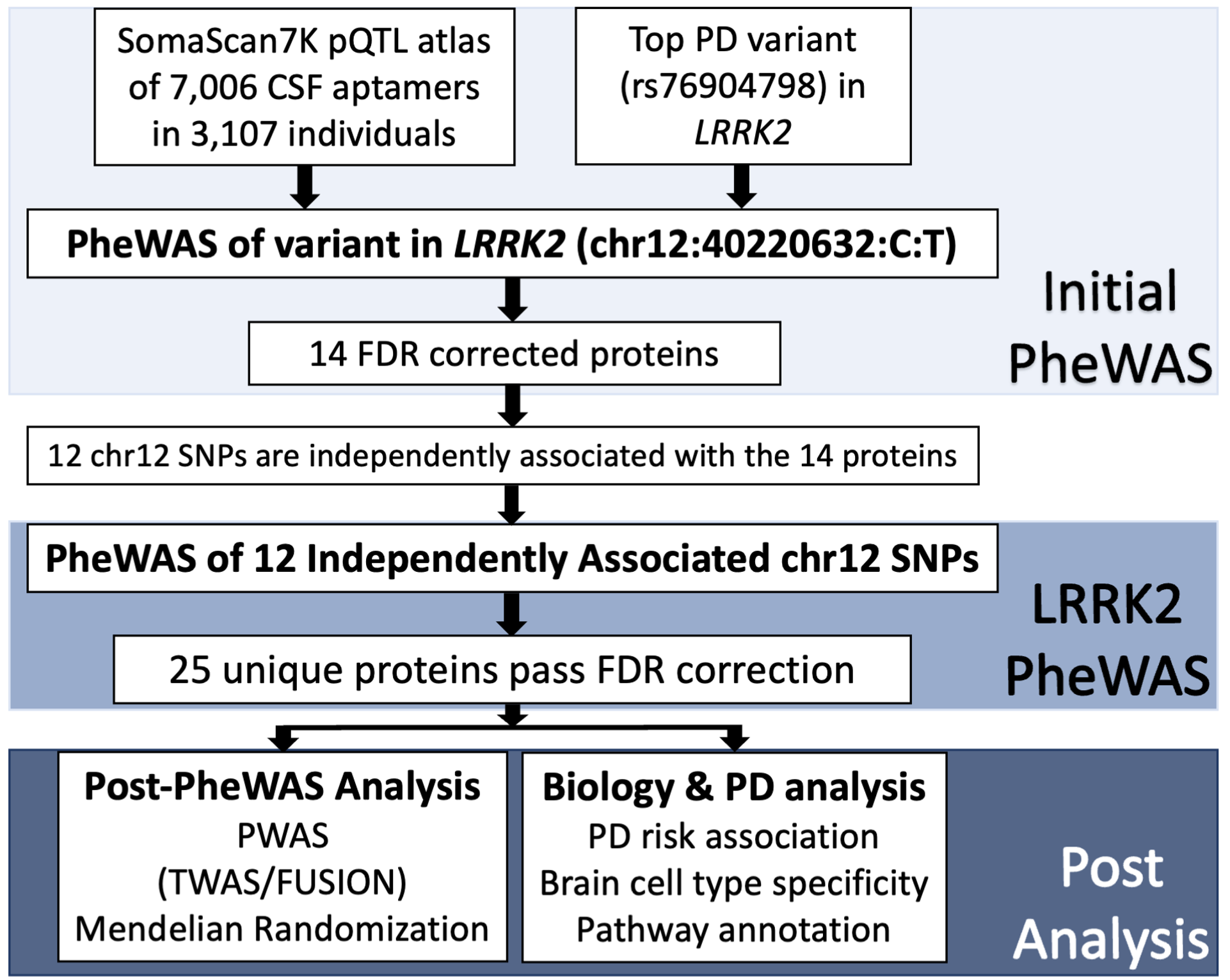
Figure 1. Study design. The workflow of the analysis of proteins associated with LRRK2 including initial rs76904798 phenome-wide association study (PheWAS), selection of independently associated LRRK2 SNPs, PheWAS of LRRK2 variants, fine mapping of significant aptamers, and analysis of proteins in terms of biological and PD significance. Adapted from Fig. 1 of the article.
We found that LRRK2 variants were associated with the CSF levels of a total of 25 proteins. Of these, 11 were previously implicated in PD based on genetic (GRN, GNMPB, CD68, HLA-DQA2, EID3, TLR3, among others) or gene expression studies (GRN, ENTPD1, CD63, GPNMB, LCT, SRI, GAA). Additionally, six other proteins (AGFG2, C1QTNF1, CHIT1, DNAJC15, FTL, OLR1) have been implicated in neurodegeneration (Alzheimer's disease (AD) and ALS) and neurological disorders (stroke)). The remaining eight proteins have not been associated with PD or neurodegeneration, although some of them (GREM2, ITGB2, SDCBP2, and TMEM106A) are known to be involved in regular neuronal function.
We used novel and powerful statistical methods to infer the proteins levels in PD cases and controls and determine that 12 proteins, including novel proteins C1QTNF1 and SDCBP2, were significant and positively associated with PD risk. Then, we used protein levels measured in prodromal, Mendelian PD cases, and controls from the Parkinson’s Progression Markers Initiative (PPMI) cohort, validating most of our predictions. Specifically, we found that HLA-DQA2, GRN, GPNMB, ENTPD1, ITBG2, and TMEM106A were dysregulated in PD cases and prodromal stages of PD, which refers to a sample listed as both a mutation carrier (LRRK2+, GBA+, or SNCA+) and control in the PPMI dataset.
Those analyses identify proteins that are dysregulated on PD, but those proteins are not necessarily part of the causal pathway. To identify causal proteins, we applied a method called Mendelian Randomization, which approximates a clinical randomized trial to identify causal proteins. Our results indicate GPNMB, CD68, LCT, and potentially ITGB2, are involved in PD pathogenesis. We then performed additional analyses to determine which biological processes and cell types these proteins are involved in and found that these proteins mainly express microglia, which is a brain cell type that regulates brain development, maintenance of neuronal networks, and injury repair. In line with those finding, these proteins are involved in leukocyte and microglial cell activation. Finally, many proteins (GPNMB, GRN, CD63, and HLA-DQA2) are part of the endolysosomal pathway also localized to the plasma membrane and had a role in immune function (Figure 2). Taken together, these results indicate LRRK2 leads to PD by disrupting the normal microglia and endolysosomal pathway and that other proteins part of these same pathways are also involved in disease pathogenesis.
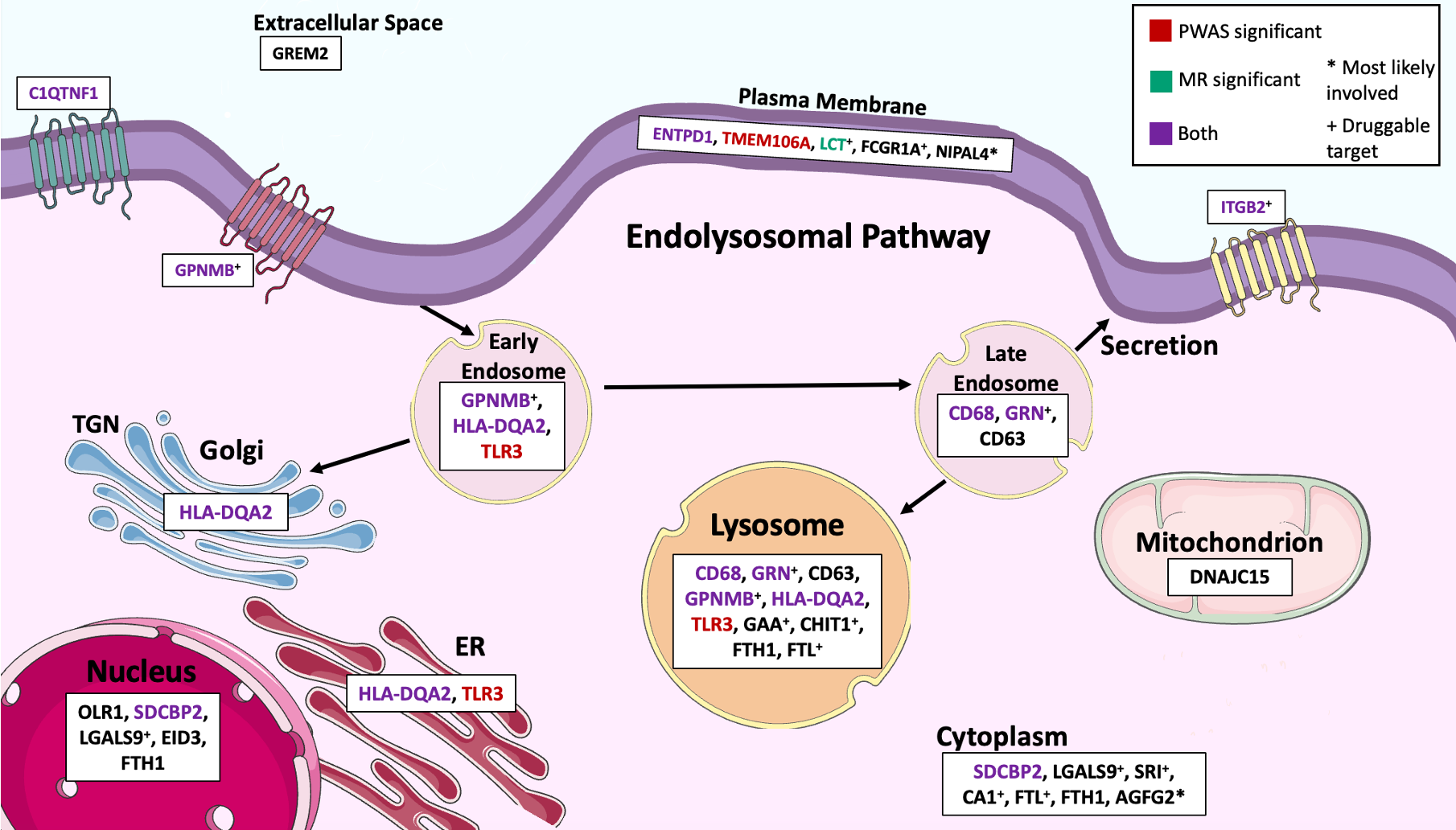
Figure 2. Cellular pathways. Pathway involvement of the 25 LRRK2 associated proteins in the endosome, Golgi apparatus, endoplasmic reticulum (ER), nucleus, mitochondrion, cytoplasm, plasma membrane, extracellular space, and lysosome. *Most likely involved. +Druggable protein target. Cell structure images are from Bioicons. Adapted from Fig. 3e of the article.
In summary, this study linked LRRK2 variants with 25 proteins, some known to be implicated in PD prior (GPNMB, GRN) and many others identified by this study for the first time (C1QTNF1 and ITGB2). CSF concentrations of proteins such as GPNMB, C1QTNF1, and ITGB2 have potential as biomarkers for target engagement and disease progression modification in clinical trials that target LRRK2. We also identified novel causal proteins that could also be novel targets for developing new PD therapies. In summary, this study is a nice example of how integrating large scale and unbiased proteomics with genetics and novel statistical approaches allows novel biomarkers, and causal and druggable targets to be identified not only for PD, but also for other neurodegenerative diseases.
References
Kim, JJ et al. Multi-ancestry genome-wide meta-analysis in Parkinson’s disease. medRxiv. 2022.
Follow the Topic
-
npj Parkinson's Disease

This journal publishes original basic science, translational and clinical research related to Parkinson's disease, including anatomy, etiology, genetics, cellular and molecular physiology, neurophysiology, epidemiology and therapeutic development and treatments.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Cognition - preclinical models, and preclinical unmet need
Publishing Model: Open Access
Deadline: Jul 27, 2026
Environmental risk factors for Parkinson’s disease
Publishing Model: Open Access
Deadline: May 13, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in