Intra- and inter-cellular recording by a 3D transistor array
Published in Electrical & Electronic Engineering
Significance
Cellular behaviors and functions rely on cell signaling. A direct approach to detect this event is to record cellular electrical potentials associated with various ionic kinetics during signal processing. It has been shown that a wide range of high-profile diseases, such as sudden cardiac arrest, cardiac arrhythmia, epilepsy, episodic ataxia, Alzheimer's, and Parkinson's diseases, may result from the dysfunction of voltage-gated ion channels. These diseases show huge impacts not only on the health-related quality of life but also on economic outcomes and place a tremendous financial burden on society. For instance, Parkinson's disease, as a chronic and progressive neurologic disorder, has affected one million people in North America. It is estimated that approximately 1% of the population over age 65 has the disease. According to the U.S. Bureau of the Census, the prevalence of PD in the United States is expected to reach 1.3 million to 1.7 million by 2040 [1]. Patients with PD have a 2.0- to 2.9-fold higher risk of death than those without the disease [2,3]. However, unfortunately, our understanding of how brain seizures originate in the brain and the underlying mechanism is lacking.
A lot of researchers have studied the electrogenic cells' electrophysiology and bridged the connections between those diseases with ion channels' activities during electrical signals orientating and propagating. The noble-prize-winning technique, the voltage clamp or patch-clamp, is such a powerful tool that can assay ion events across those voltage-sensitive ion channels and 'clamp' the cell membrane potential to aimed values realizing the manipulation of ion activities. By doing so, we have accumulated numerous insights on cellular physiological activities; therefore, we could develop therapies and approaches to combat relevant diseases. However, although qualitative knowledge of ions' motions has been well studied, quantitative understanding is still missing because of the lack of tools that allow high-spatiotemporal-resolution sampling inside the cells.
Design and Approach
To this end, we seek to develop more delicate and advanced electrophysiological tools for biologists and facilitate understanding the dynamic activity of cellular circuits.
We need to consider four grand challenges to engineering an effective electronic interface. First, the interface must penetrate the cell membrane and obtain a strong signal inside the membrane. Conventional electronic bioprobes, such as a multi-electrode array [4] and a multi-transistor array [5], are planar, large, and rigid, limiting their ability to access the inner side of the cell membrane. As a result, the signal recording would lose one-to-one correspondence, signal magnitude, and quality [6-8]. Second, the internalization of the interfaces must show minimal invasiveness to the cells. Glass pipettes used in patch-clamp require rupturing the cell membrane during signal recording. Nano-sized bioprobes can easily penetrate the cell membrane and form giga-seal. A soft device would reduce the mechanical mismatch between the interface and cell, so have reduced invasiveness and high stability during recording. Third, the interface needs to be accurate. Passive nanowire-based conductive electrodes usually exhibit significantly attenuated signals due to their high electrical impedances, limiting their ability to pick up low magnitude sub-threshold cell membrane potentials. Fourth, the device must be scalable. Studying the electrical properties in a cellular network requires simultaneous interfacing of multiple cells, thus, it requires the interface has more than one sensing site.
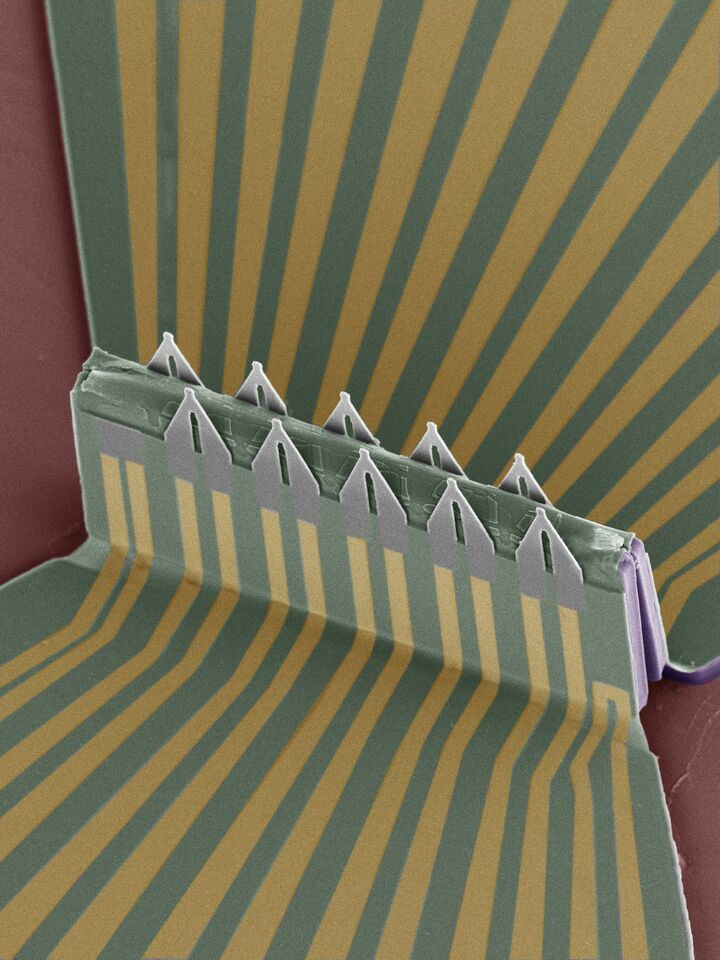
We addressed these challenges with a 3D field-effect transistor (FET) array- stretchable, scalable, high-sensitivity- engineered using micro/nano-fabrication techniques and the compressive buckling technique. The array shows four appealing features. First, we design the 3D geometry of the sensors with fine tips picking up signals inside the cell body (Figure 1). The FET's tip is approximately 1~2 μm wide, small enough to insert into HL-1 and mouse cardiac muscle cells (~50 µm).
Second, we coat the FET surface with a phospholipid bilayer to facilitate the insertion process. The lipids used in this work include extracted cell membranes of red blood cells and a synthetic lipid, DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine). With this functionalized coating, the FET would access the cytosol after the spontaneous fusion of the phospholipid and the cell membrane (Figure 2).
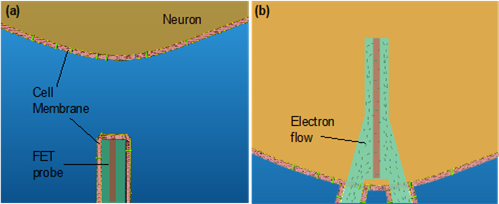
internalization into the target cell. The phospholipid facilitates the insertion
process, enhancing the tight sealing at the interface and also reducing the mechanical
invasiveness of the probe.
Third, the FET sensor can actively sense the intracellular events with an amplitude as large as those by the gold-standard patch-clamp. The tiny part on the probe that is intracellular interfacing with the cell in the active sensor's gate terminal, which could sense the electrical potential variation around itself, and converts the gate potential change to a conductance difference of the FET. Owing to this working principle, the FETs would exclude the influence of high interfacial electrical impedance allowing sensing of subthreshold potentials.

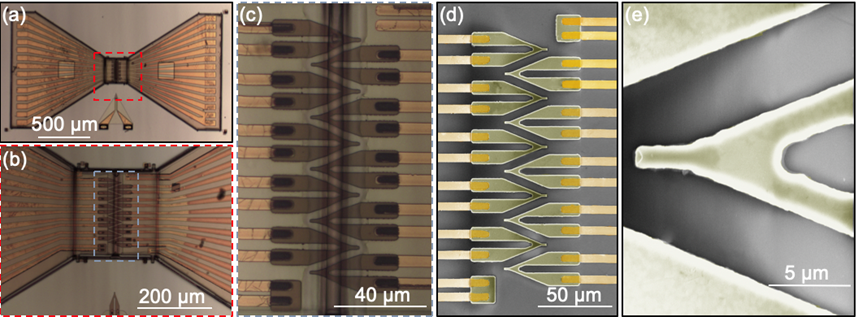
and (b) the compressive buckling of 2D functional precursors to fabricate the 3D FET array
on a pre-strained elastomeric substrate.
Forth, we can boost the number of sensors (e.i. FETs) to realize large-scale cell recording. The soft array was fabricated by the compressive buckling technique, an approach that was successfully demonstrated by Sheng Xu lab at UCSD [9]. In Figure 3, we illustrated the fabrication process of the 3D FET array.
Application
We used the FET array to measure electrophysiological signals of single cells, both intracellularly and extracellularly, and compared the shape and magnitude of these signals to those by the patch-clamp and multi-electrode array. The similarities in these comparable recordings verify the cells are in the proper state and validate the 3D FET array can faithfully record the electrophysiological signals. Additionally, we sensed the sub-threshold signals (~15 mV in magnitude) from the intracellular action potential recordings. Drug screening is a vital function of electrophysiological tools, so we used the FET sensor to monitor the action potential change after administering ion-blocking drugs to the cells. The FET can accurately record the corresponding responses to the drugs.
Inter-cellular information flow plays a central role in electrophysiological regulation that governs activities on the single-cell level and also on the network level, such as during microcircuit formation, memory readout, and decision making [10]. Simultaneously sensing and recording these signals in between the cells with single-cell precision will provide insights into these aspects. We studied the propagative behaviors of electrical signals in the cardiac muscle network using the 10-FET array on 2D cell cultures, and the 128-FET array on 3D cell cultures. Intracellular action potentials are recorded from the FETs. The signal conduction velocity was calculated from the simultaneous recordings of different cells. Notably, we directly recorded the signal propagation in a cell, for the first time, and discovered that the intracellular conduction velocity is ~5 times that of the intercellular conduction. The results are exciting because they show the potentials of using the FET array for signal propagation researches of electrogenic cells and obtaining vital insights related to diseases cause by false signal conduction inside one cell or between neighboured cells. Intracellular recording of cells in 3D cell cultures is challenging because the sensor-cell interface is difficult to form. By utilizing the phospholipid coating on the FET surface, a stable intracellular FET-cell interface can be formed stably; thus, intracellular action potentials were successfully recorded, for the first time. The success of sensing intracellular electrophysiology of a 3D cell culture paves the way to record signals of the brain or heart in vivo, which would provide pivotal biological insights into brain or heart functions.
For more details, please see the original version of the manuscript in Nature Nanotechnology:
https://www.nature.com/articles/s41565-021-01040-w
Acknowledgments
The author thanks Prof. Sheng Xu for his precious advice and careful revisions.
References
[1] S. L. Kowal, T. M. Dall, R. Chakrabarti, M. V. Storm, A. Jain, The current and projected economic burden of Parkinson's disease in the United States, Movement Disorders 28, 311-318 (2013) https://onlinelibrary.wiley.com/doi/abs/10.1002/mds.25292
[2] D. A. Bennett, L. A. Beckett, A. M. Murray, K. M. Shannon, C. G. Goetz, D. M. Pilgrim, D. A. Evans Prevalence of Parkinsonian Signs and Associated Mortality in a Community Population of Older People, New England Journal of Medicine 334, 71-76 (1996) http://www.nejm.org/doi/full/10.1056/NEJM199601113340202
[3] M. M. Hoehn, M. D. Yahr, Parkinsonism: onset, progression, and mortality, Neurology 50, 318-318 (1998) https://www.ncbi.nlm.nih.gov/pubmed/9484345
[4] J. Pine, Recording action potentials from cultured neurons with extracellular microcircuit electrodes, Journal of Neuroscience Methods 2, 19-31 (1980) http://www.sciencedirect.com/science/article/pii/0165027080900424
[5] A. Lambacher, M. Jenkner, M. Merz, B. Eversmann, R. A. Kaul, F. Hofmann, R. Thewes, P. Fromherz, Electrical imaging of neuronal activity by multi-transistor-array (MTA) recording at 7.8 mu m resolution, Applied Physics a-Materials Science & Processing 79, 1607-1611 (2004) http://link.springer.com/article/10.1007%2Fs00339-004-2991-5
[6] W. Lu, C. M. Lieber, Nanoelectronics from the bottom up, Nature Materials 6, 841-850 (2007) http://dx.doi.org/10.1038/nmat2028
[7] T. Cohen-Karni, B. P. Timko, L. E. Weiss, C. M. Lieber, Flexible electrical recording from cells using nanowire transistor arrays, Proceedings of the National Academy of Sciences 106, 7309-7313 (2009) http://www.pnas.org/content/106/18/7309.abstract
[8] J. F. Eschermann, R. Stockmann, M. Hueske, X. T. Vu, S. Ingebrandt, A. Offenhäusser, Action potentials of HL-1 cells recorded with silicon nanowire transistors, Applied Physics Letters 95, 083703 (2009) https://doi.org/10.1063/1.3194138
[9] S. Xu, Z. Yan, K.-I. Jang, W. Huang, H. Fu, J. Kim, Z. Wei, M. Flavin, J. McCracken, R. Wang, A. Badea, Y. Liu, D. Xiao, G. Zhou, J. Lee, H. U. Chung, H. Cheng, W. Ren, A. Banks, X. Li, U. Paik, R. G. Nuzzo, Y. Huang, Y. Zhang, J. A. Rogers, Assembly of micro/nanomaterials into complex, three-dimensional architectures by compressive buckling, Science 347, 154-159 (2015) http://science.sciencemag.org/content/sci/347/6218/154.full.pdf
[10] L. A. Jorgenson, W. T. Newsome, D. J. Anderson, C. I. Bargmann, E. N. Brown, K. Deisseroth, J. P. Donoghue, K. L. Hudson, G. S. F. Ling, P. R. MacLeish, E. Marder, R. A. Normann, J. R. Sanes, M. J. Schnitzer, T. J. Sejnowski, D. W. Tank, R. Y. Tsien, K. Ugurbil, J. C. Wingfield, The BRAIN Initiative: developing technology to catalyse neuroscience discovery, Philosophical Transactions of the Royal Society of London B: Biological Sciences 370, (2015) http://rstb.royalsocietypublishing.org/content/royptb/370/1668/20140164.full.pdf
Follow the Topic
-
Nature Nanotechnology

An interdisciplinary journal that publishes papers of the highest quality and significance in all areas of nanoscience and nanotechnology.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in