Investigating Maternal Autoantibodies and their Impact on Offspring Neurodevelopment
Published in Neuroscience and General & Internal Medicine
Autism and Maternal Autoantibodies
Autism is a widely studied and highly heterogeneous neurodevelopmental condition. As awareness of autism has increased, so too has the potential for earlier diagnoses and implementation of therapeutic interventions. However, understanding its causes remains an active area of research. To date, there are several known etiologies for the development of autism, and a high incidence of idiopathic cases still exists.
Maternal autoantibody-related autism (MARA) is a subtype of autism linked to the presence of specific patterns of autoantibodies (MARA-ABs) in maternal circulation. Clinical studies identified that the targets for MARA-ABs are proteins known to be important for neurodevelopment. Our work herein expands upon previous research to elucidate how the known clinical patterns of MARA-ABs alter offspring neurodevelopment and ultimately cause this subtype of autism. Developing a better understanding of this autism subtype and the MARA-ABs will allow us to identify therapeutics that protect the developing brain against MARA-ABs.
A New Iteration of the MAR Rat Model
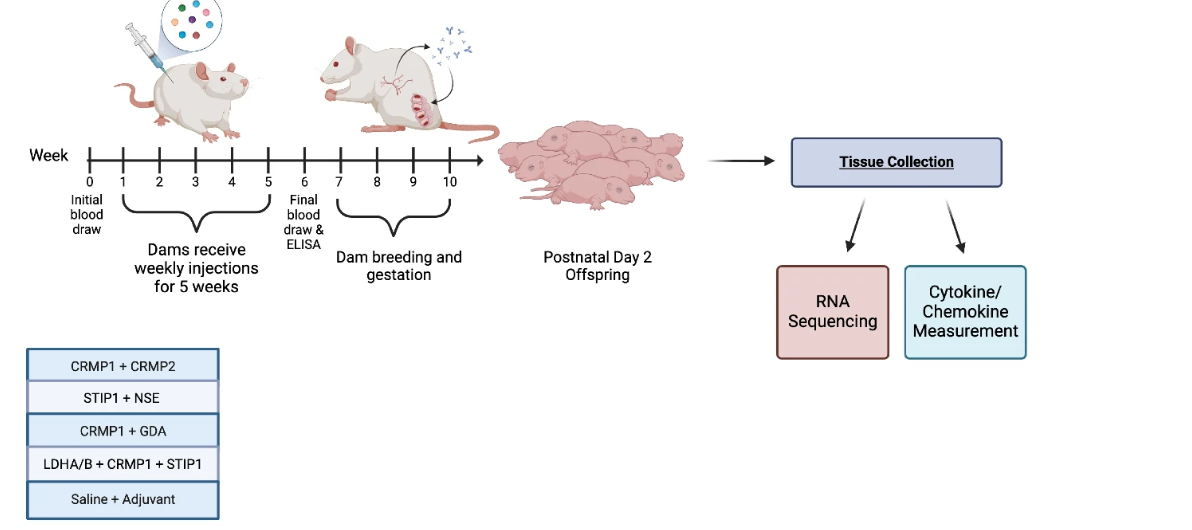
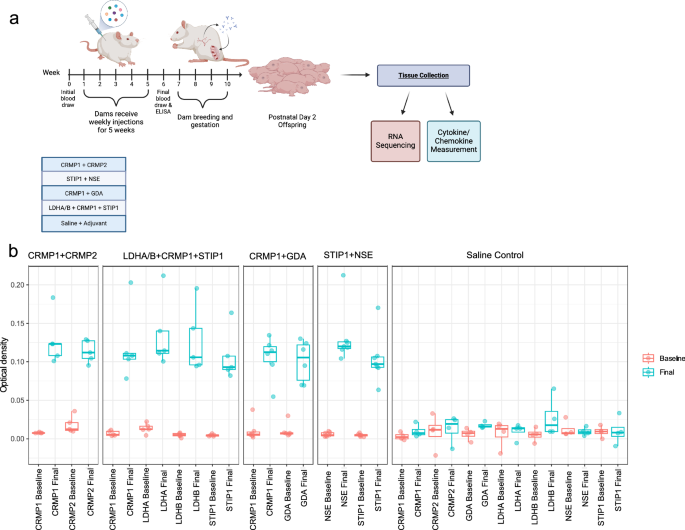
Our lab has utilized several iterations of mouse and rat models to better understand how gestational autoantibody exposure impacts offspring neurodevelopment. Using our rat model, previous studies identified behavioral and neuroanatomical changes as a result of exposure to one pattern of MARA-ABs: collapsin response mediator protein 1 (CRMP1) and stress-induced phosphoprotein 1 (STIP1) and lactate dehydrogenase A and B (LDHA/B). However, following the completion of that study, we have identified additional clinical patterns of MAR-ABs that are not only highly correlated with autism, but we found that some are also associated with a more severe autism phenotype.
In our current study, we immunized rat dams such that they produced autoantibodies to the following combinations of proteins: CRMP1+CRMP2, STIP1 and neuron-specific enolase (NSE), CRMP1 and guanine deaminase (GDA), as well as the original LDHA/B+CRMP1+STIP1. Once immunized, the rat dams produced autoantibodies continuously, even during pregnancy. As a result, offspring are exposed throughout gestation as would occur clinically. This model allows us to assess the effects of the autoantibodies in the absence of any other environmental or immune perturbations that could also cause neurodevelopmental changes.
Using postnatal day 2 offspring from the immunized dams, we assessed changes in the brain transcriptomics and in the brain and peripheral immune signaling molecules.
Genetic and Immune Impacts of MARA-ABs
When we assessed the offspring from dams immunized against the various MARA-AB patterns, we found that there were impacts on gene expression within the brain, and changes to the levels of cytokines and chemokines in the brain and in the periphery. Globally, we found that exposure to any of the MARA-ABs results in changes to the numbers of up and downregulated genes. However, based on MARA-AB pattern, we found that those offspring gestationally exposed to the CRMP1+CRMP2 pattern of MARA-ABs were the most profoundly affected. When we examined some of the specific genes altered, we noted upregulation of Wnt signaling genes and downregulation of immune signaling genes, including those that induce NFkB. These changes could contribute to the development of several cell types within the brain, brain overgrowth, and have downstream consequences on long term learning and memory (Figure 1).
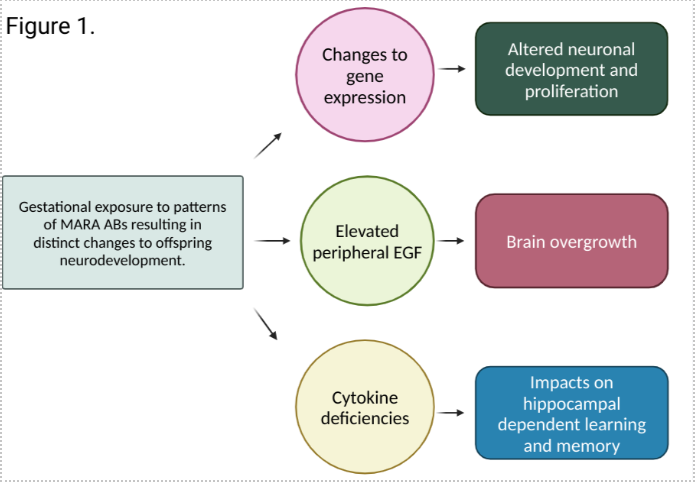
The genetic changes that we observed were supported by higher levels of epidermal growth factor, and lower levels of several key cytokines, chemokines, and growth factors in the brain that play roles in the development of neural cells within key brain regions that have been implicated in autism, including the hippocampus. Further, we noted that based on the MARA-AB exposure, additional growth factors such as vascular endothelial growth factor and interleukin-2 were differentially affected.
The full details of our results are published in the article entitled “Gestational Autoantibody Exposure Impacts Early Brain Development in a Rat Model of MAR Autism.”
The MAR-ASD Phenotype
From this work, we confirmed two main points: 1) the impacts of gestational MARA-AB exposure can be detected at the genetic and cell signaling levels; 2) there are differential impacts on early neurodevelopment based on the pattern of MARA-AB exposure. We found that in the offspring exposed to the MARA-ABs, changes to the gene expression and cytokines and chemokines suggested a phenotype consistent with brain overgrowth and alterations in the cells required for proper learning and memory. These findings mirror what has been observed clinically in MARA cases, including the highest impact being related to exposure to antibodies against CRMP1+CRMP2.
Future Directions
The latest iteration of our rat model yielded several insightful findings. However, several questions still remain. How do these autoantibodies continue to impact the brain and offspring behavior throughout their lifetime? Studies to address the varying impacts of the different MARA-AB patterns longitudinally are underway. In addition, the mechanisms by which the MARA-ABs interact with their cellular targets remain unclear. Studies to understand how the ABs gain access to their protein targets and the consequences of their interactions are also in progress.
MARA is a unique subtype of autism that is linked to the maternal production of specific ABs. While our studies continue to broaden our understanding of how these ABs impact the neurodevelopmental trajectories of the offspring, our ultimate goal is to develop therapeutic strategies to protect the offspring brain from the ABs during the vulnerable and vital periods of development.
Follow the Topic
-
Molecular Psychiatry

This journal publishes work aimed at elucidating biological mechanisms underlying psychiatric disorders and their treatment, with emphasis on studies at the interface of pre-clinical and clinical research.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in