Investigating the hidden genomic background of complex diseases: the story of iSNP, the integrated single nucleotide polymorphism network platform
Published in Healthcare & Nursing

Ulcerative Colitis is one of the most common types of inflammatory bowel disease. The current understanding of ulcerative colitis pathogenesis suggests genetic predisposition and environmental triggers combine to lead to continuous inflammation in the gut. Sadly there is no real cure, only various levels of treatments. Some patients can be kept in equilibrium with mesalazine treatment; others require surgery to avoid dangerous flare-ups and a trip to intensive care.
Our questions were centred around this disease. Can we find some common mechanisms behind these varying phenotypes? Do we have one disease or multiple? To answer the questions we developed our investigator: the integrated Single nucleotide polymorphism Network Platform - iSNP.
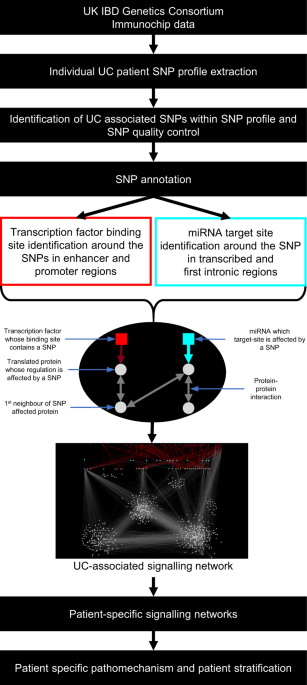
The iSNP workflow begins with a patient’s single nucleotide polymorphism data, which might be taken from an immunochip. Then, iSNP filters the SNPs known to be involved in ulcerative colitis, thanks to the herculean effort of previous genome-wide association studies. These SNPs are checked as to whether they are in regulatory (non-coding) regions of the genome, i.e. enhancers, promoter areas, or miRNA target sites. If an SNP alters a transcription factor binding site in an enhancer or promoter region or a miRNA target site then the iSNP method annotates the corresponding protein as an SNP-affected protein. Next, it checks the interactors of the SNP-affected protein using the OmniPath protein interaction network, similar to what we did in a study focusing on cancer pathomechanism. In this way, iSNP creates a regulatory network signature of each patient with ulcerative colitis.
These network signatures told us that stress response, NFKB signalling and cytokine production were the most commonly involved pathways in ulcerative colitis. These are the usual suspects in ulcerative colitis, but we found that how they are reached is different from patient to patient. The network signatures in the various patients can be separated into four clusters by the high degree proteins. The affected pathways include calcium signalling and angiogenesis through VEGFA. This suggests that multiple pathways lead to the same outcome in ulcerative colitis.
But of course, if iSNP found something in one modality that could just be an artefact, so we validated our genomic and network-based results using an orthogonal transcriptomic approach. We downloaded the largest ulcerative colitis dataset from the TAMMA database. Here we found most of the SNP-affected genes are differentially expressed, into two clusters out of our four. We also found the usual suspects amongst the differentially expressed genes. Based on this, we can say that our iSNP investigator was successful, in that it identified the usual suspects like immune functions and the NFKB1 pathway, but also found something interesting such as the involvement of VEGF and calcium signalling in ulcerative colitis.
But we won’t celebrate the complete success of iSNP until we can use it on a large enough dataset, which we weren’t able to do to date. Like many others, we are looking forward to seeing a larger dataset, like the one created in the UK IBD Bioresource.
As in complex conditions, there are many pathways where our investigation could go now such as using non-long coding RNAs for predicting the effect of SNPs, or using a tissue-specific network. However, we all hope that you could check out iSNP and maybe you can also use it in your project to find the pathway to understanding complex diseases.
Meet iSNP here: https://github.com/korcsmarosgroup/iSNP
In the name of the authors:
Jo, Dezso and Tamas
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in