Large-scale single cell sequencing uncovers the epididymis cell atlas and mitochondria segmental-speicific distribution
Published in Healthcare & Nursing

The epididymis is crucial to sperm maturation. Spermatozoa produced from testis acquire their capacity of motility and fertilizing ability during epididymal transit. Multiple studies (including our own here) have reported different genes in the epididymis and facilitated sperm maturation. However, the exact cell composition and gene expression repertoire of each cell population in the epididymis are less characterized. Several research groups including ourselves have tried to elucidate the cell compositions of the epididymis via recently hot technique-single-cell sequencing (scRNA-seq) at a similar time.
While cell atlas has been identified by single-cell sequencing (scRNA-seq) 1,2, this work, as far as we know, represents the first to report the spatiotemporal differences of segmental difference expression genes (DEGs) and mitochondria along the epididymis with sufficient cell number. Here, we have demonstrated a total of eight-cell types by a total of 40,623 cells scRNA-seq. Benefiting from our high-throughput scRNA-seq, we have identified a subpopulation of principal cells with stereocilia at gene expression level that was not described in the other studies (Figure 1). 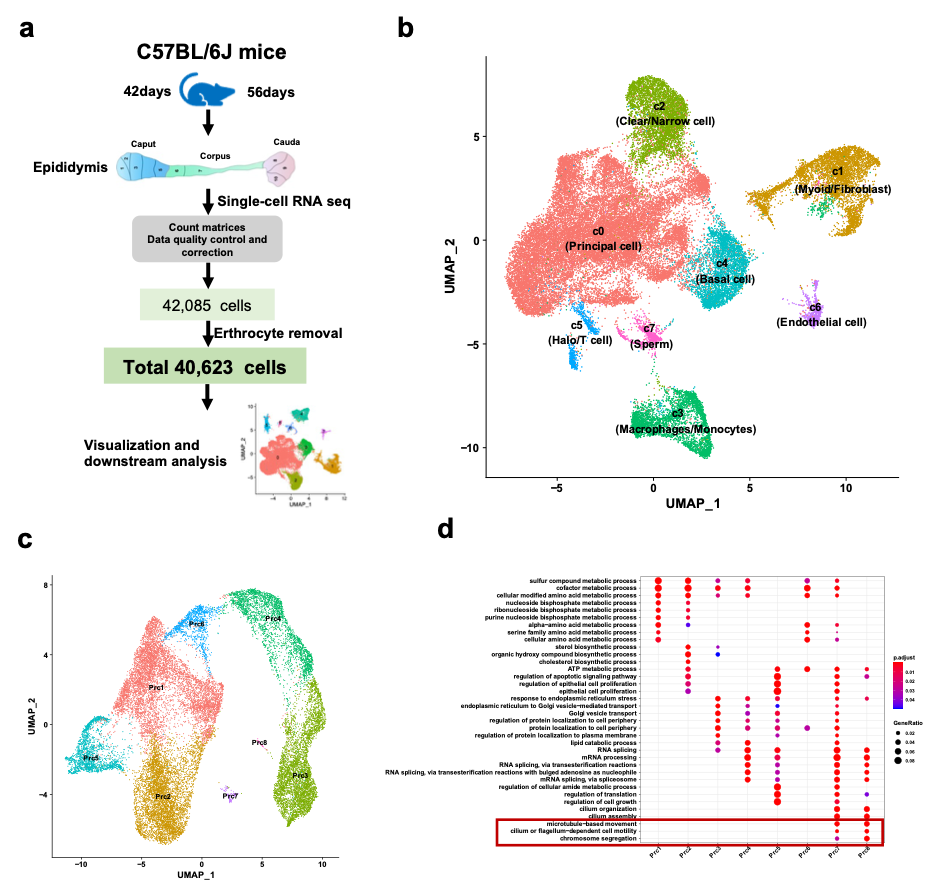
Figure 1. Overview of single-cell RNA sequencing of mouse epididymis regions and principal cell subpopulations.
Unexpectedly, our results revealed the high content of mitochondria in the corpus and cauda epididymis compared with the caput using scRNA-seq. Although we noticed the high percentage of mitochondrial transcriptions (MTs%) during the analysis, we did not pay a lot of attention to this issue when we first submitted the manuscript. We just thought the phenomorphan was accounted to one of mitochondria-rich apical cells in the caput epididymis. During peer review, two referees questioned our high percentage of MTs% due to the MTs% of corpus epididymis over 20%. In general, a high percentage of MTs (>5%) is considered an indicator of cell stress or cell death 3. To address the referees’ concerns which definitely is a major issue in the analysis of scRNA-seq, we sent more samples to supplement our previous data. Consistent with our previous results, a similar MT% was observed. Additional experiments including mitochondrial specific staining and qPCR validation revealed a higher number of mitochondria and mitochondrial gene expressions in the corpus and cauda epididymis (Figure 2). These results strongly suggest that the high MT% in our data was NOT due to the low-quality cells but attributed to the high percentage of mitochondria in the corpus and cauda epididymis.
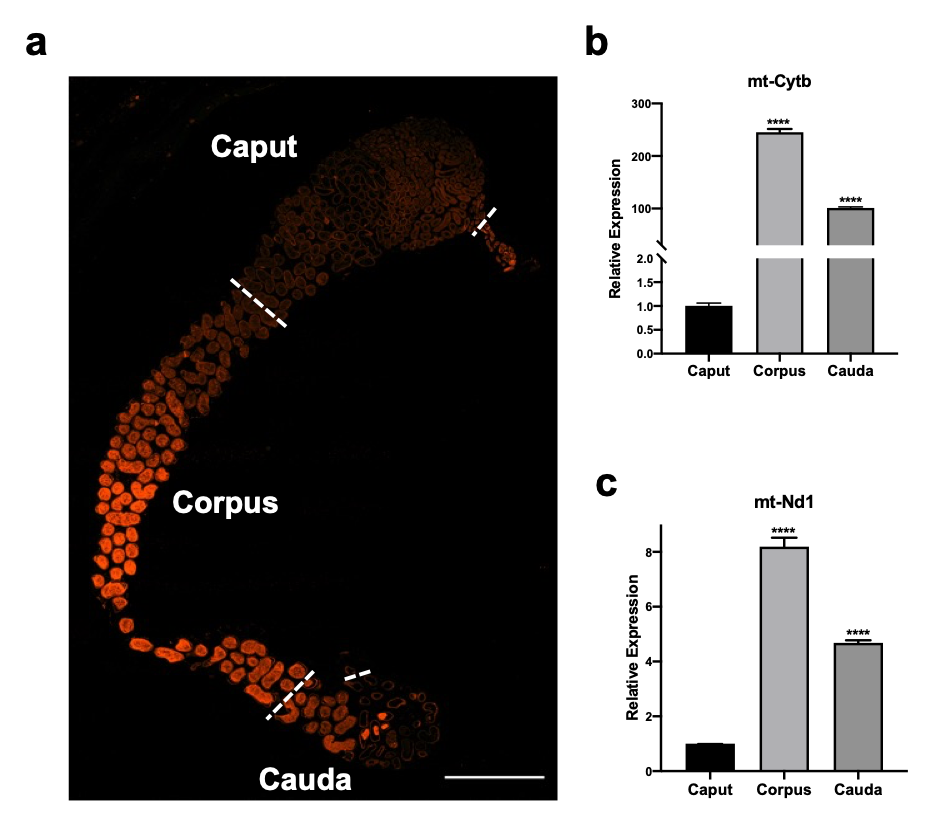
Figure 2. The segmental specific distribution of the mitochondrial in the epididymis.
Taken together, our data provide an indispensable resource, at single-cell resolution, for a comprehensive cell composition of the epididymis and revealed the novel distributed pattern of mitochondria and key genes that could be linked to the sperm functionalities in the first wave and subsequent wave of sperm.
For more details, please find our paper with supplementary data on Cell Discovery (https://www.nature.com/articles/s41421-021-00260-7).
References
1 Leir, S. H., Yin, S., Kerschner, J. L., Cosme, W. & Harris, A. An atlas of human proximal epididymis reveals cell-specific functions and distinct roles for CFTR. Life Sci Alliance 3, doi:10.26508/lsa.202000744 (2020).
2 Rinaldi, V. D. et al. An atlas of cell types in the mouse epididymis and vas deferens. Elife 9, doi:10.7554/eLife.55474 (2020).
3 Ilicic, T. et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17, 29, doi:10.1186/s13059-016-0888-1 (2016).
Follow the Topic
-
Cell Discovery

This journal aims to provide an open access platform for scientists to publish their outstanding original works and publishes results of high significance and broad interest in all areas of molecular and cell biology.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in