LOV2FRET - a simple method to optimise and validate illumination trains for cellular optogenetics
Published in Bioengineering & Biotechnology

Repurposing naturally occurring photosensor domains to control biochemical pathways is sometimes known as cellular optogenetics and has greatly expanded the horizons of biology. The substantial information already known about protein interactions can potentially now be harnessed to provide precise spatiotemporal regulation of virtually any biological process. We were initially inspired by demonstrations of light-controlled actin regulation in cell-free (1), and subsequently cell-based (2,3), systems to embark on a project using known binding motifs to regulate MAPKs. We aimed to investigate the significance of the timing of JNK signaling (4,5) as this could not be addressed by conventional reverse genetics (6).
Building the required synthetic proteins exhibiting target binding in cell-free conditions was relatively straightforward (7). However, demonstrating an optical actuator has the intended dynamically switchable functions in intact cells presented a major bottleneck. First, MAPKs do not necessarily cause rapid and visible translocation of their endogenous targets, so assays to demonstrate function can be slow and indirect. Secondly, white light illumination is cytotoxic in the long term, limiting the duration of experiments (8). The illumination should be limited, for example by using a pulsed blue-light source, but still sufficient to activate the photosensor. Determining the required amount of light is complicated not only by the indirectness of functional readouts but also by spontaneous relaxation of photosensors to an inactive state. This may be one reason that cellular optogenetics has to some extent focused on regulated translocation to and from the nucleus, plasma membrane, mitochondrial membrane or other compartments (e.g. 9,10,11).
These are extremely valuable approaches for many applications, but there are nevertheless cases where an optogenetic equivalent of a spatiotemporally controlled small molecule, regulating a target at a chosen time or place but not necessarily causing instant visible changes, would also be useful. As phototoxicity limited the duration of our optogenetic experiments (8), greater actuator sensitivity would be beneficial. For every new construct, how do we determine the light needed to switch the photosensor in living cells, so that we do not apply more light than needed? How often should we provide more light to compensate for the spontaneous inactivation of the sensor i.e. how do we determine the inactivation rate in intact cells?
We had been tagging new blue-light actuator designs to cyan fluorescent proteins to allow multiplexing with green, red and infrared reporter systems. We realized that resonance energy transfer (RET) between the fluorescent protein tag and the photosensor chromophore is likely, but were concerned that it was unknown whether this would have appreciable effect on actuator function. To address this question, we repurposed the elegant light-driven nuclear exporter LEXY (9) to our needs. We found that replacing the traditional red tag with a cyan one increased photon sensitivity 7-fold (12). Increasing the distance between tag and photosensor domain eliminated this difference, implying that RET has a substantial effect on actuation sensitivity (12). Moreover, as energy can be lost from either the tag or the photosensor, RET makes the fluorescent tag either dimmer or brighter. As the photosensor is photochromic, the quenching or dequenching changes dynamically as switching takes place. Therefore the already common practice of fluorescently tagging photosensors, at least in the case of the popular blue-light activated LOV2 domain from Avena sativa, can address all three difficulties presented by non-translocating actuators – limited sensitivity, lack of in situ visual readout for switch activation and spontaneous relaxation – depending on the tag used (Fig. 1, 12).
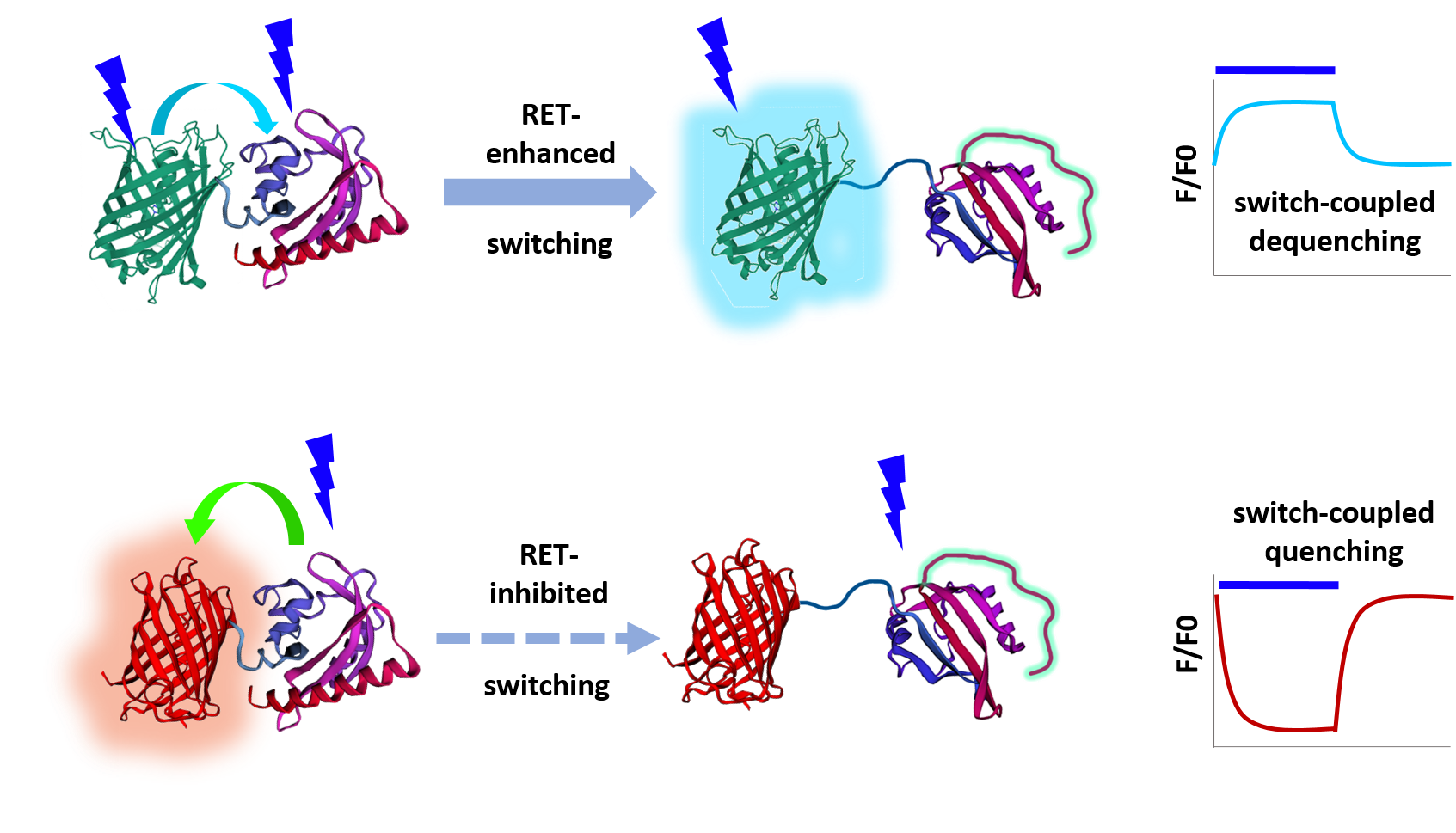
RET-based resolution of these three issues makes temporal patterning of pathways much more straightforward because we can now precisely compute the predicted dynamics of an optogenetic switch in response to any illumination train. We can now reduce JNK activation with the original MAPK inhibitor design optoJNKi using extremely low light levels (12). We can determine which relaxation mutant photosensor is best matched to reshaping a signaling pathway profile, giving a 10-fold improvement in throughput. This, in turn, allows rapid estimation of kinetic constants of pathway activation/inhibition and how these change during evolution of response. Moreover, it is possible to dynamically alter the photosensitivity of an actuator by using a reversibly photoswitchable protein (12). This concept could in the future allow multiplexing of individually addressable blue-light actuator species in the same cell, using alternate wavelengths or other cues to specify which species is sensitive to blue light pulses at any given time.
References
- Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc Natl Acad Sci U S A. 2008 Sep 2;105(35):12797-802. doi: 10.1073/pnas.0801232105. Epub 2008 Aug 26. PMID: 18728185; PMCID: PMC2525560.
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009 Sep 3;461(7260):104-8. doi: 10.1038/nature08241. Epub 2009 Aug 19. PMID: 19693014; PMCID: PMC2766670.
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009 Oct 15;461(7266):997-1001. doi: 10.1038/nature08446. Epub 2009 Sep 13. PMID: 19749742; PMCID: PMC2989900.
- Coffey ET, Smiciene G, Hongisto V, Cao J, Brecht S, Herdegen T, Courtney MJ. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci. 2002 Jun 1;22(11):4335-45. doi: 10.1523/JNEUROSCI.22-11-04335.2002. PMID: 12040039; PMCID: PMC6758801.
- Cao J, Semenova MM, Solovyan VT, Han J, Coffey ET, Courtney MJ. Distinct requirements for p38alpha and c-Jun N-terminal kinase stress-activated protein kinases in different forms of apoptotic neuronal death. J Biol Chem. 2004 Aug 20;279(34):35903-13. doi: 10.1074/jbc.M402353200. Epub 2004 Jun 10. PMID: 15192112.
- Björkblom B, Vainio JC, Hongisto V, Herdegen T, Courtney MJ, Coffey ET. All JNKs can kill, but nuclear localization is critical for neuronal death. J Biol Chem. 2008 Jul 11;283(28):19704-13. doi: 10.1074/jbc.M707744200. Epub 2008 May 12. PMID: 18474608.
- Melero R, Tuittila M, Yadav L, Min J, Courtney MJ (2013) Design and development of optogenetic tools to manipulate signalling pathways with high spatiotemporal precision in neuronal cells. SFN 2013-S-16331.
- Melero-Fernandez de Mera RM, Li LL, Popinigis A, Cisek K, Tuittila M, Yadav L, Serva A, Courtney MJ. A simple optogenetic MAPK inhibitor design reveals resonance between transcription-regulating circuitry and temporally-encoded inputs. Nat Commun. 2017 May 12;8:15017. doi: 10.1038/ncomms15017. PMID: 28497795; PMCID: PMC5437309.
- Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B. Optogenetic control of nuclear protein export. Nat Commun. 2016 Feb 8;7:10624. doi: 10.1038/ncomms10624. PMID: 26853913; PMCID: PMC4748110.
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012 Mar 4;9(4):379-84. doi: 10.1038/nmeth.1904. PMID: 22388287; PMCID: PMC3444151.
- Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods. 2016 Sep;13(9):755-8. doi: 10.1038/nmeth.3926. Epub 2016 Jul 18. PMID: 27427858; PMCID: PMC5137947.
- Li LL, Klein FM Li Greci L, Popinigis A, Freudenberg F, Courtney MJ Resonance energy transfer sensitizes and monitors in situ switching of LOV2-based optogenetic actuators. Nat. Commun. 2020 Oct 9;11(1):5107. doi: 10.1038/s41467-020-18816-8. PMID: 33037199.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in