Mapping the human cortex in vivo with diffusion MRI
Published in Neuroscience and Protocols & Methods
In their 1993 perspective, "The Backwardness of Human Neuroanatomy," Crick and Jones called for technologies to map the microarchitecture of the living human brain. While diffusion MRI (dMRI) has since led to remarkable progress in mapping white matter, our in-vivo understanding of cerebral cortical microstructure has lagged. Research has largely been limited to macroscale measurements or a few myeloarchitectural features. Our paper, "Mapping the Microstructure of Human Cerebral Cortex In Vivo with Diffusion MRI," aims to address this gap by generating comprehensive neuromaps of cortical microstructure from dMRI and investigating their relationship to the brain's molecular, cellular, functional, and electrophysiological organization.
Distilling Cortical Microstructure: From 21 Metrics to Four Factors
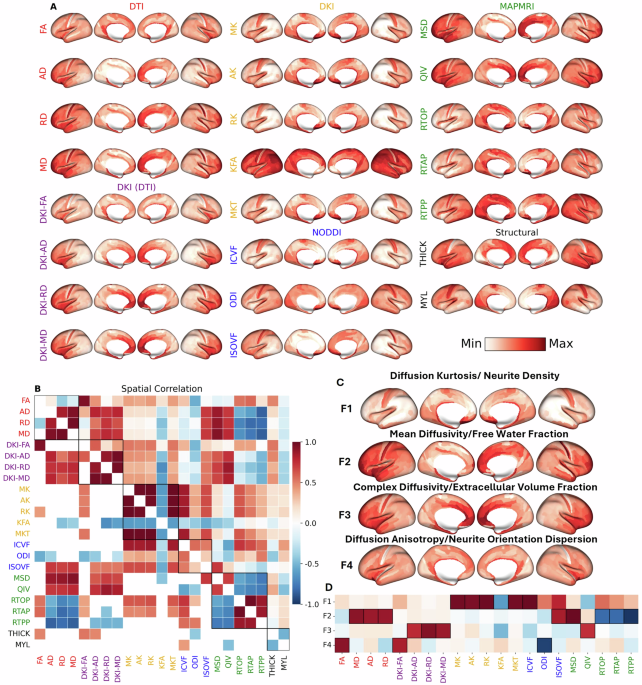
Leveraging high-quality data from the Human Connectome Project (HCP), we derived 21 microstructural metrics from the cerebral cortex of 962 healthy adults. These metrics were computed from several common dMRI signal representations and tissue models: Diffusion Tensor Imaging (DTI), Diffusion Kurtosis Imaging (DKI), Mean Apparent Propagator (MAP-MRI), and Neurite Orientation Dispersion and Density Imaging (NODDI).
In the first version of the manuscript, we used t-distributed stochastic neighbor embedding (t-SNE) to display the inter-relatedness of the dMRI metrics. While t-SNE is commonly used for dimensionality reduction, in our case it lacked sufficient interpretability and in hindsight probably wasn’t the best tool for the job. We received several comments from peer reviewers complaining about the complexity of 21 dMRI metrics and their difficulty interpreting the results. While the metrics used are well known throughout the dMRI community, to those less familiar, dealing with 21 metrics from four signal representations/tissue models must have felt daunting. Those interested in this initial version can check out the first version of our bioRxiv preprint. Thanks to an anonymous reviewer, who helpfully suggested using factor analysis, we were able to
- Rewrite our manuscript to not require in-depth knowledge of dMRI models.
- Distill our results and figures to focus on the most pertinent aspects of the dMRI metrics.
When presenting our work at the 2025 annual meeting of the International Society for Magnetic Resonance in Medicine (ISMRM) in Hawaii, we ran into Rafael Henriques-Neto. He pointed us to his own concurrent work conducting factor analysis in white matter dMRI which we were unaware of. From this experience, we realized that
- The field badly needed and probably still needs more work to go into interpreting the now vast array of signal representations/signal models in dMRI.
- Even today in the era of social media, informal chats at in-person conferences are essential for keeping research communities well-informed and connected.
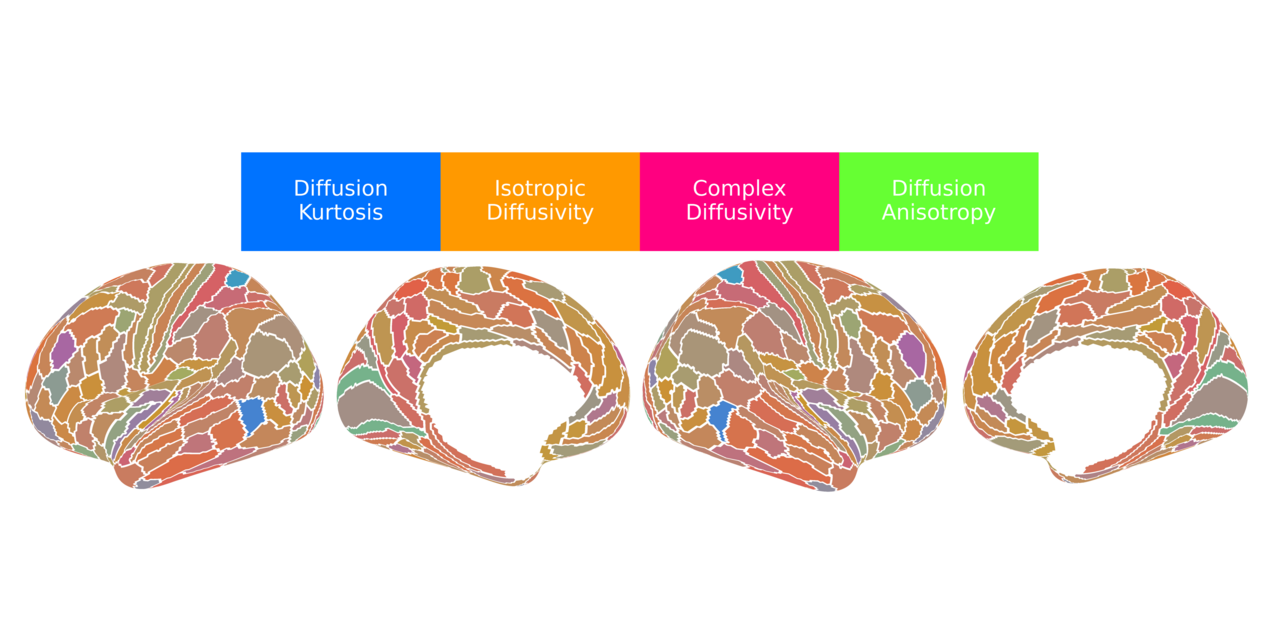
Our application of dimensionality reduction via factor analysis revealed nearly 90% of the variance across the 21 dMRI metrics could be explained by four underlying, independent factors that have both a physical basis (given first) and a biological interpretation (given second):
- Factor 1 (F1) - Diffusion Kurtosis/Intracellular Volume Fraction: This factor was most heavily loaded by metrics such as mean, axial, and radial kurtosis (MK, AK, RK) and was positively correlated with the intracellular volume fraction (ICVF) from NODDI, which corresponds to neurite density. Shown in blue, this factor is most concentrated in primary sensorimotor cortices and lowest in heteromodal association areas, particularly within the prefrontal and lateral parietal lobes.
- Factor 2 (F2) - Isotropic Diffusivity/Free Water Fraction: This factor corresponded to the isotropic volume fraction (ISOVF) or free water fraction from NODDI and was positively correlated with DTI-derived diffusivities and the mean-squared displacement (MSD) from MAP-MRI. Shown in orange, this factor was most concentrated in high-order association cortices, particularly the prefrontal cortex, anterior temporal lobe, and the insula. It was lowest in primary and secondary visual areas and medial sensorimotor regions.
- Factor 3 (F3) - Complex Diffusivity/Extracellular Volume Fraction: This factor captured diffusion in heterogeneous microenvironments, anchored by the quantitative inverse variance (QIV) from MAP-MRI and positively correlated with DKI-derived diffusivities, likely reflecting the extracellular volume fraction. Shown in magenta, this factor followed a pattern similar to F2, with the highest values in the anterior cingulate, medial prefrontal cortex, and insula. It showed the lowest values in primary sensorimotor and most visual cortices. Interestingly, F3 was strongly correlated with the principal gradient of gene expression among the neuromaps that define the sensorimotor-association (SA) axis, suggesting that alterations of gene expression are required for complex tissue microenvironments in the cerebral cortex.
- Factor 4 (F4) - Diffusion Anisotropy/Neurite Orientation Dispersion: This factor was loaded by fractional anisotropy (FA, DKI-FA) and was negatively correlated with the neurite orientation dispersion index (ODI) from NODDI. Shown in lime green, this factor was highest in the insula and limbic/paralimbic areas and lowest in primary sensory areas.
Importantly, we were able to replicate these four composite factors in an independent dataset and also got similar factors from Independent Component Analysis (ICA). We were also surprised to find the cortical dMRI metrics had high test-retest repeatability, demonstrating the feasibility of in-vivo gray matter dMRI.
Advancing In-Vivo Neuroanatomy
In this publication, with valuable feedback from anonymous peer reviewers and other colleagues, we have created a more tractable framework for studying the cerebral cortex in vivo. Artificial Intelligence (AI) foundation models run on clusters of GPUs with identical (or near-identical) internal architectures; however, the human brain is wired very differently. The gray matter of the cerebral cortex is organized into many different regions with diverse microarchitectures, each specialized for efficient processing of vital tasks ranging from sensory perception (visual, auditory, tactile, proprioceptive, vestibular, etc.) to high-level reasoning and judgement to emotional regulation. Our dMRI findings show that this cortical microstructure is systematically aligned with cytoarchitectural and laminar hierarchies, macroscale functional gradients, electrophysiological dynamics, neurotransmitter systems, and cognition/behavior. The next step is to extend this work by constructing brain networks of gray matter regions with similar internal architectures using diffusion MRI, leveraging the principle of “homophily”, that regions sharing structural similarities are likely to be connected and work together in coordination. In doing so, this line of research helps resolve the "backwardness of human neuroanatomy" by integrating a rich new layer of dMRI-derived information into the multimodal neuromaps framework, advancing our understanding of how the brain's intricate structure gives rise to its complex functions.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in