Measuring the impact of chromatin context on transcription factor binding affinities
Published in Bioengineering & Biotechnology
Transcription factor (TF) binding across the genome is involved in regulating numerous biological processes and is often dysregulated in disease. However, because TF binding depends on a complex interplay between TF abundances, DNA sequence, and chromatin context, when and why TFs bind to specific subsets of their potential binding sites is not well understood1,2.
In our previous work on intestinal organoid differentiation, we found that binding of some TFs that regulate differentiation increased to a point at which these TFs were no longer restricted to known and expected binding sites3. Instead, binding occurred at all accessible regulatory elements, which confused our motif and target-gene overrepresentation analyses. Expression analysis then revealed that protein abundances sometimes reached millions of molecules per cell, presumably saturating all accessible regulatory elements in the DNA. This result made us realize that it is essential to consider TF abundance and the epigenetic landscape when studying TF biology; however, there were no tools available to measure these factors in the native chromatin context.
In recent years, various sequencing-based methods that enable charting the epigenetic landscape and transcription factor binding across the genome have been developed4,5. However, determining the concentration at which a TF binds to chromatinized DNA — the binding affinity — was previously not possible at a genome-wide scale. This complicates the interpretation and integration of epigenetic and gene expression data, as it is difficult to predict the consequences of changes to either the epigenome or TF abundance on TF binding and subsequent gene expression regulation.
To determine TF binding affinities to chromatinized DNA, we developed Binding Affinities to Native Chromatin by sequencing (BANC-seq), enabling us to measure the concentration at which a TF can bind its potential binding sites in the genome and how this is influenced by the epigenetic landscape. In BANC-seq, isolated nuclei are incubated with a titration series of a FLAG-tagged TF. TF concentration-dependent binding is then measured across the genome using chromatin immunoprecipitation followed by sequencing (ChIP-seq) or cleavage under targets and release using nuclease (CUT&RUN) to sequence bound DNA. Binding affinities are then computed for each binding site by fitting a Hill equation binding curve to the binding signal across TF concentrations (Fig. 1a). Because the epigenetic landscape is kept intact during TF binding, the detected binding affinities depend both on the local chromatin context and the underlying DNA sequence. Therefore, BANC-seq data is uniquely suited to investigate how chromatin context impacts TF binding and affinities and to predict how changes in TF abundance result in changes in TF binding across the genome.
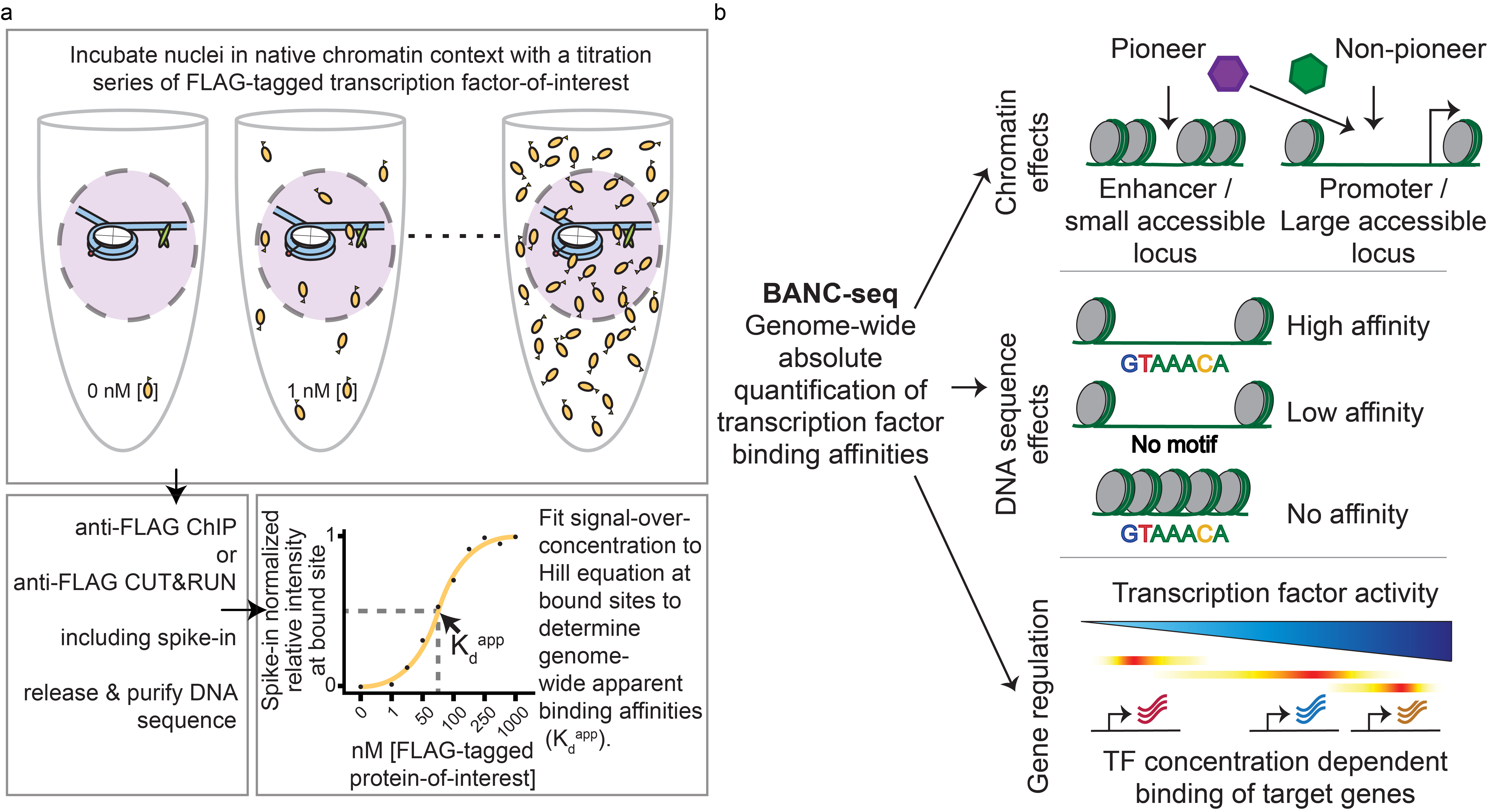
Figure 1 | Overview of the BANC-seq experimental procedure and interpretation. a. Nuclei are incubated with a concentration range of a FLAG-tagged TF, followed by ChIP-seq or CUT&RUN with spike-in controls (for normalization), and computation of absolute apparent TF binding affinities (KdApps) at each binding site by fitting normalized sequencing reads to a Hill equation binding curve. b. Illustration of how TF binding affinity is regulated by chromatin context, DNA sequence, and the concentration-dependent binding of TFs to their targets.
Performing BANC-seq for the TFs YY1, MYC/MAX, SP1, and FOXA1 revealed thousands of nanomolar-affinity binding sites for each TF. By integrating our BANC-seq data with epigenome data, we found that the chromatin context of a binding site, especially its DNA accessibility, is a major determinant of nanomolar binding affinities. Surprisingly, high-affinity binding sites were mostly found at promoter regions, indicating that these sites are the first to be occupied at low TF expression levels. However, FOXA1 seemed to be an exception as it was less restricted by pre-existing accessibility or the presence of promoter elements, possibly because of its role as a pioneer TF which enables it to directly bind condensed chromatin. Furthermore, BANC-se
q revealed that DNA sequence motifs and a permissive chromatin environment work in concert to facilitate TF binding, where near-perfect consensus motifs are especially important for high-affinity binding but are less important for lower-affinity binding (Fig. 1b).
BANC-seq enables us to investigate the interplay between the epigenome, DNA sequence and TF abundance to be quantified. This new type of quantitative data enables the stratification of TF binding sites by their required TF concentration, given a certain epigenetic state, and has implications when analysing TF binding motifs and regulatory networks. These analyses can now be extended to incorporate TF concentration-dependent motifs and target genes. Furthermore, BANC-seq aids the interpretation of biological events that are characterized by changes in TF abundance, such as the overexpression of reprogramming factors or disease-associated (onco)genes.
It is important to note that although BANC-seq can give insights into the TF binding potential in a particular, static epigenetic environment, it does not evaluate changes in binding affinities that may lie downstream of TF binding and the subsequent remodelling of the epigenome. In addition, BANC-seq requires tagged, recombinant TFs that can be purified at a suitable concentration while remaining biologically active, which potentially complicates experiments with poorly soluble TFs and restricts the testable concentration range.
Future efforts will include extending the amount of affinity resolved TF binding profiles across TF families and epigenetic states using BANC-seq in conjunction with matched epigenome profiling. We hope that BANC-seq will provide insights into how TFs dynamically interact with the epigenome, other TFs and chromatin remodellers, allowing us to uncover how the epigenome is read and written by TFs.
References
- Guo, J., et al. (2014). "Sequence specificity incompletely defines the genome-wide occupancy of Myc." Genome Biology 15(10): 482.
- Slattery, M., et al. (2014). "Absence of a simple code: how transcription factors read the genome." Trends Biochem Sci 39(9): 381-399.
- Lindeboom, R. G., et al. (2018). "Integrative multi-omics analysis of intestinal organoid differentiation." Mol Syst Biol 14(6): e8227.
- Dunham, I., et al. (2012). "An integrated encyclopedia of DNA elements in the human genome." Nature 489(7414): 57-74.
- Lambert, S. A., et al. (2018). "The Human Transcription Factors." Cell 172(4): 650-665.
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in