Mettl15 promotes the maturation of mitochondrial ribosomes and facilitates cell lactylation by regulating the posttranslation of 12S rRNA
Published in Cell & Molecular Biology
Mitochondria play a crucial role as a primary provider of ATP, serving as indispensable components in the cellular energy production pathway known as oxidative phosphorylation (OXPHOS). When mitochondria experience dysfunction, it disrupts OXPHOS activity, resulting in compromised cellular energy metabolism, heightened levels of reactive oxygen species (ROS), increased oxidative stress, and ultimately, cell death. These mitochondrial-related issues are closely associated with aging, diabetes, neurodegenerative diseases, cardiovascular disorders, and various forms of cancer. Understanding and addressing mitochondrial function is pivotal in mitigating the impact of these health challenges.
Mammalian mitochondria harbor a distinctive ~16.5 kb circular DNA genome, transferring the genetic instructions for 2 mitochondrial ribosomal RNAs (12S and 16S), 22 mitochondrially-encoded transfer RNAs, and 13 protein constituents integral to the oxidative phosphorylation (OXPHOS) system. The translation of these 13 protein component genes is orchestrated by mitochondrial ribosomes (mitoribosomes). Mitoribosomes consist of the 28S mitochondrial small subunit (mt-SSU), housing the 12S rRNA and 30 nucleus-encoded mitoribosomal proteins, and the 39S mitochondrial large subunit (mt-LSU), containing the 16S rRNA and 52 nucleus-encoded mitoribosomal proteins. Functionally crucial regions of the mitoribosome contain several post-transcriptional modifications, pivotal in mitoribosome assembly and efficient translation. Disruptions in the expression of mitochondrial rRNA modification enzymes directly impact modification levels of mt-rRNA and mitoribosome assembly, culminating in impaired mitochondrial function and the onset of various mitochondrial diseases. Understanding and addressing these intricacies are paramount for unraveling the complexities of mitochondrial-related disorders.
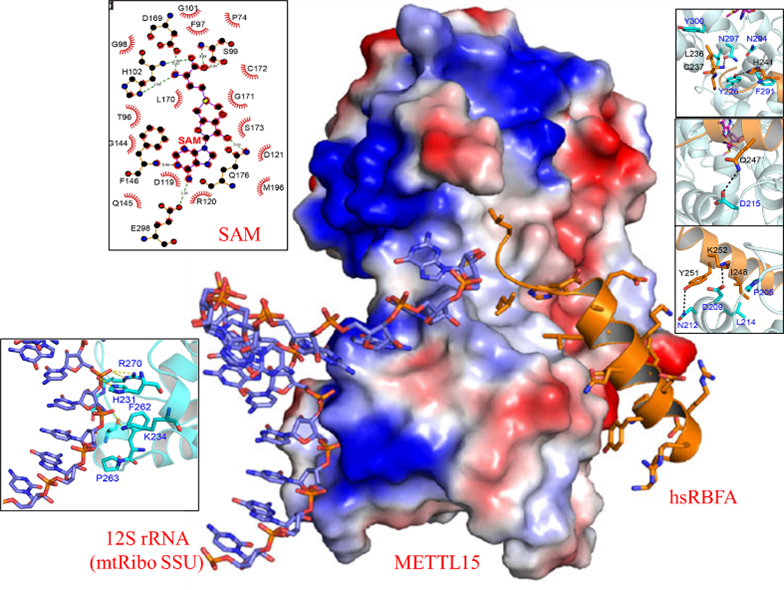
The Methyltransferase-like (METTL) family proteins exhibit a distinctive conserved Rossman-like fold S-adenosyl methionine (SAM)-binding domain, enabling them to methylate a range of substrates, including proteins, nucleic acids, and small molecule metabolites. These proteins play a crucial role in regulating mRNA stability and translation efficiency. Among them, the N4-methylcytidine (m4C) methyltransferase METTL15 has been identified as the critical factor responsible for the specific recognition and modification of m4C839 in the 44th cervical ring structure of the mitochondrial small subunit (mt-SSU) 12S rRNA (named as helix 44). The inactivation of the human METTL15 gene (or its mouse homolog, Mettl15) has been linked to disturbances in the translation of mitochondrial protein-coding mRNAs, leading to a decrease in mitochondrial respiration capacity. Notably, research has indicated a potential correlation between METTL15 and childhood obesity. Additionally, mitoribosome assembly and stability, crucial for mitochondrial function, rely on ribosome-binding factors, such as hsRBFA. As a scaffold protein, hsRBFA is recruited to mt-SSU 12S rRNA helix 44 and helix 45, facilitating proper decoration, assembly, and maturation of mt-SSU.Recent studies highlight that hsRBFA undergoes a significant conformational change, ensuring the completion of TFB1M-mediated dimethylation of helix 45, followed by the recruitment of METTL15 for further modification. Our group's previous research has demonstrated that hsRBFA employs a novel binding mode to recognize 12S rRNA. This binding depends on its KH-like domain and the basic amino acids in its N-terminus. This dual dependence effectively promotes the proper assembly of mitoribosomes, shedding light on the intricate regulatory mechanisms governing mitochondrial translation and function.
To unravel the intricate molecular interactions among METTL15, hsRBFA, and 12S rRNA, and to delineate the regulatory role of METTL15 in mitochondrial gene expression, our study conducted an in-depth analysis of a serial of crystal structures, including the structures of the methyltransferase METTL15 apo form, METTL15 in complex with its methyl donor S-adenosyl methionine (SAM), the ternary complex of METTL15-SAM-hsRBFA, and the quaternary complex of METTL15-SAM-hsRBFA-12S rRNA. Structural analysis revealed that METTL15 comprises the Methyltransferase (MTase) domain and a scaffold-like domain, which can recognize the methyl donor SAM through relatively conserved pockets. Simultaneously, hsRBFA was observed to bind to the hydrophobic surface on one side of the METTL15 scaffold-like domain via its C-terminal helix. Notably, the substrate RNA was found to bind to the opposite side of the scaffold-like domain. Additional experimental techniques, including mutagenesis, isothermal titration calorimetry (ITC), and fluorescence polarization (FP), were employed to identify critical residues crucial for the specific recognition of METTL15 by hsRBFA and RNA. These findings suggest that hsRBFA plays a pivotal role in recruiting METTL15 to mitochondrial ribosomal small subunits, emphasizing its significance in the process rather than direct binding to 12S rRNA (Fig. 1). This comprehensive structural and experimental analysis provides valuable insights into the molecular mechanisms underlying the interaction between METTL15, hsRBFA, and 12S rRNA, shedding light on their collective roles in the regulation of mitochondrial gene expression.
Moreover, we observed that the knockdown of METTL15 in 293T cells exerts a profound impact not only on the translation levels of the 13 proteins encoded by mitoribosomes but also on mitochondrial oxidative phosphorylation (OXPHOS) activity and cellular metabolic pathways. Specifically, this knockdown resulted in notable alterations such as a significant increase in mitochondrial reactive oxygen species (ROS) levels, a decrease in mitochondrial membrane potential, a reduction in mitochondrial tricarboxylic acid cycle levels, and a compensatory increase in glycolysis levels. Comprehensive analyses using non-targeted metabolomics and Western blot (WB) techniques further unveiled that METTL15 knockdown in cell lines induces an elevation in anaerobic respiration levels. This shift towards anaerobic respiration was associated with an upregulation of intracellular lactate secretion levels. Notably, these metabolic changes were linked to alterations in histone lactylation modifications in the nuclear genome, with a specific upregulation of histone H4K12-la and H3K9-la modifications (Fig.2). These findings underscore the multifaceted role of METTL15 in cellular physiology, revealing its influence not only on mitochondrial protein translation and OXPHOS activity but also on broader cellular metabolic pathways and histone modifications. The study provides valuable insights into the intricate regulatory network orchestrated by METTL15 and its implications for cellular homeostasis.
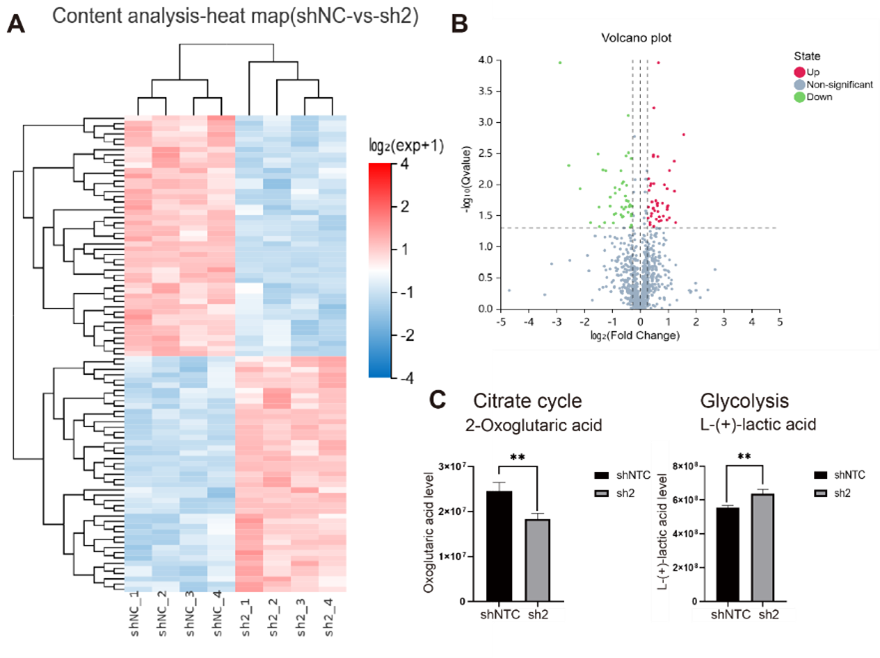
Figure 2. Knockdown of METTL15 leads to changes in cellular metabolic pathways and increased lactate levels
This study employed a multifaceted approach, integrating structural biology, biochemistry, and cell biology techniques to thoroughly and systematically analyze the recognition mechanism between the methyltransferase METTL15 and its diverse substrates. By delving into the consequences of METTL15 deficiency, the research elucidated its intricate impact on mitochondrial function and cellular metabolism. Additionally, the study explored how mitochondrial metabolites influence the epigenetic regulation of the nuclear genome. These comprehensive findings contribute novel insights into the previously underexplored epigenetic regulatory role of METTL15 in mitochondrial gene expression. Furthermore, the study serves as a valuable reference for subsequent investigations into the structure and function of other methyltransferases within mitochondria. The integrated approach applied in this research advances our understanding of METTL15's multifaceted functions. It paves the way for broader exploration into the complex interplay between mitochondrial biology and nuclear epigenetic regulation.
Follow the Topic
-
Cell Discovery

This journal aims to provide an open access platform for scientists to publish their outstanding original works and publishes results of high significance and broad interest in all areas of molecular and cell biology.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcement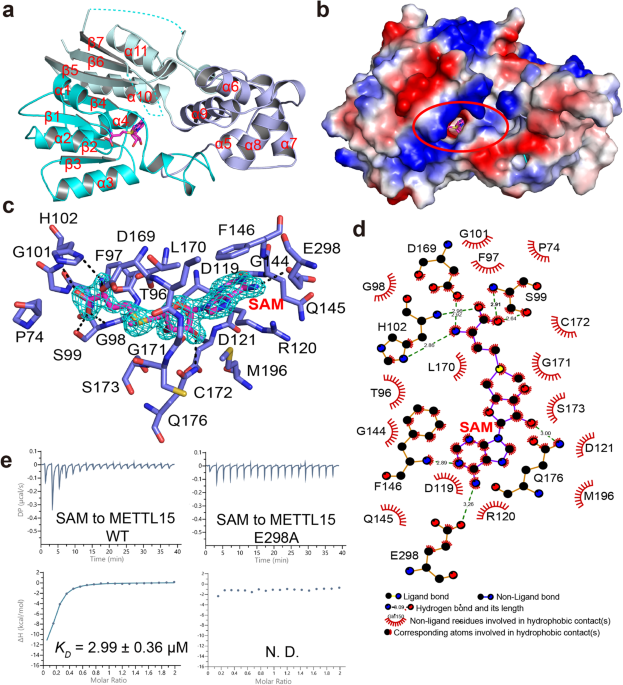
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in