Missing parent of a deadly yeast pathogen found… in the sea!
Published in Microbiology

Most people associate the word yeast with wine, beer, or bread. For centuries, we humans have been taking advantage of the fermenting abilities of some yeasts for our own benefit, but not all yeasts are so kind to us. In fact, there is a group of diverse yeasts—named Candida—that can thrive inside our bodies, cause disease and even turn into deadly pathogens, particularly when our defences are weak. There are over 30 different species of Candida that can infect humans and most of them are rarely, but increasingly, found and because of that, we call them “emerging yeast pathogens”. But, from where do they emerge? This is a difficult question that has puzzled us for many years. Back in 2014, our group found that some clinical strains of one such emerging species, Candida orthopsilosis, were hybrids between two parental lineages diverging around 5% at the nucleotide level (Pryszcz et. al. 2014). Since then, we and others have shown that many other rare Candida pathogens were also of a hybrid nature. Even the established opportunistic pathogen Candida albicans seems to descend from an ancient hybridisation event (Mixo and Gabaldón 2020). This places hybridisation as a potential mechanism to generate new strains with pathogenic potential (Mixo and Gabaldón 2018).
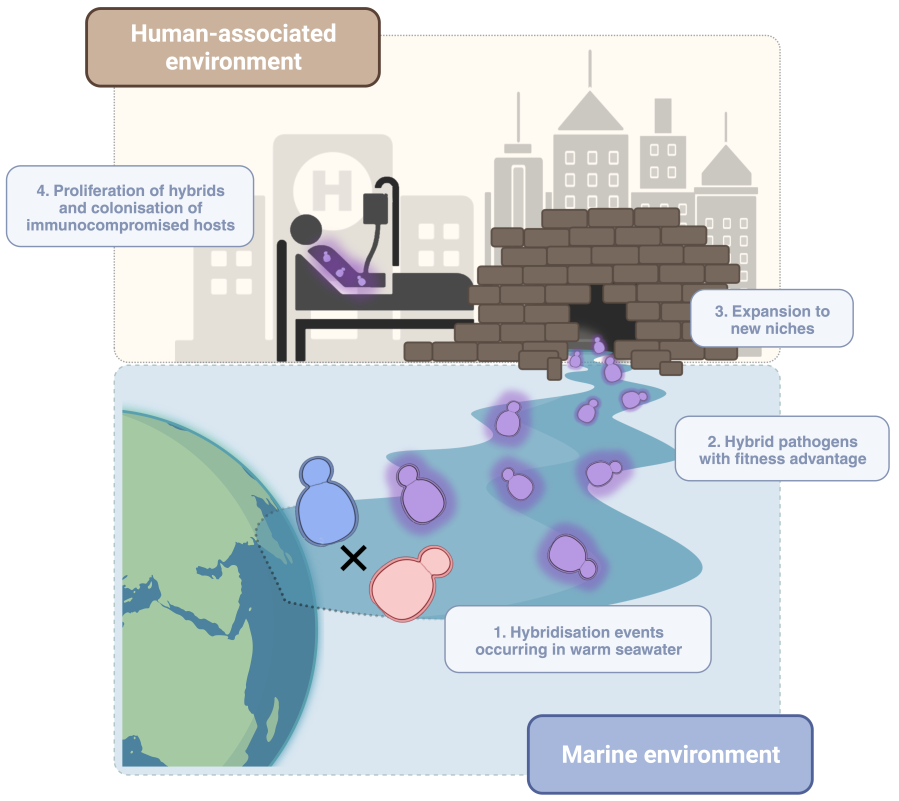
The case of C. orthopsilosis remained a paradigmatic one. We and others sequenced many other clinical strains, which revealed that hybrids between the same two parentals had been formed multiple independent times, and all resulting hybrid clades had become globally distributed. Although most of the strains were hybrids, one of the homozygous parental species—we called it parent A—could be rarely found in the clinics too, but we’d never seen its soulmate (parent B). One problem here was that we only had access to clinical samples, and the missing parent could be either non-virulent or live in an unknown environment. We started a search for C. orthopsilosis strains isolated from the environment. In the literature we found that, based on ITS identification, this species was often isolated from marine samples. We corresponded with authors of such publications and requested the strains, often without success. The strains were not kept, could not be sent, or our request was left without a response. We finally connected with a colleague that was part of a Dutch/Qatari study of the yeast diversity on coastal environments in Qatar. They had also found this species there, and kindly shared the strains with us.
We sequenced them and found that, although most of the marine strains were hybrids with highly heterozygous genomes, there were a couple of very homozygous samples which corresponded to parental strains. We quickly realised that these new marine parentals were quite different to the known parent A. After a series of comparative and population genomics analyses we came to the conclusion that they corresponded to the alternative parental lineage, which together with parental A, had given rise to all C. orthopsilosis hybrids known to date. So there it was, we were finally looking at the elusive parent B. This unexpected and exciting finding allowed us to have a complete overview of the genome evolution of C. orthopsilosis and ask questions like: which traits do hybrids inherit from each of their parentals? Why are hybrids much more commonly found than parental lineages? Are parents A and B different at a phenotypic level too? Why hasn’t parent B been found in the clinics? Is it an avirulent strain? For several months, our group worked hard to find answers to all these questions. We performed studies at a genomic as well at a phenotypic level and obtained some interesting results. What we found was that, at a genomic level, the hybrids inherited about half of their genomic material from each parent, without apparent strong selection on the regions that were inherited or on the parental of which they came from. However, things took a twist when we studied their phenotype and ability to grow under adverse conditions. One of our most interesting findings was to discover that the newly-found parent B grew at high temperatures (38 to 42ºC), an ability that parent A lacked completely and that hybrids had inherited to some extent. This is a very important trait, one crucial for pathogenesis, as our first barrier of defence against fungal infections is our basal body temperature. The tolerance to antifungal drugs seemed to also be intrinsically different amongst parentals and inherited by hybrids to different degrees. To our surprise, we found that parent B and marine hybrids were as virulent as strains isolated from the clinics. It was also interesting to see that hybrid strains isolated from the clinics or from the marine environment did not differ much at genomic or phenotypic level. This, which might seem unimportant at first, suggests an environmental origin of this species and tells us that these pathogens likely did not evolve to specifically infect us. But instead, those features enabling them to survive in the marine environment might also be useful for proliferating inside the human body. Now, if we take all these results together, it seems that hybridisation, which we hypothesise could be happening in warm marine waters, could play a key role in the emergence of new hybrid pathogens. We might then want to factor in climate change, and the more and more recurrent episodes of extreme heat, which could be acting upon these strains with intrinsic pathogenic potential. This could lead to the selection of thermotolerant strains capable of breaching our thermal barrier and causing disease, especially when our immune systems—our second defence barrier—are debilitated.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in